|
chr1_-_169703178
|
3.445
|
NM_000450
|
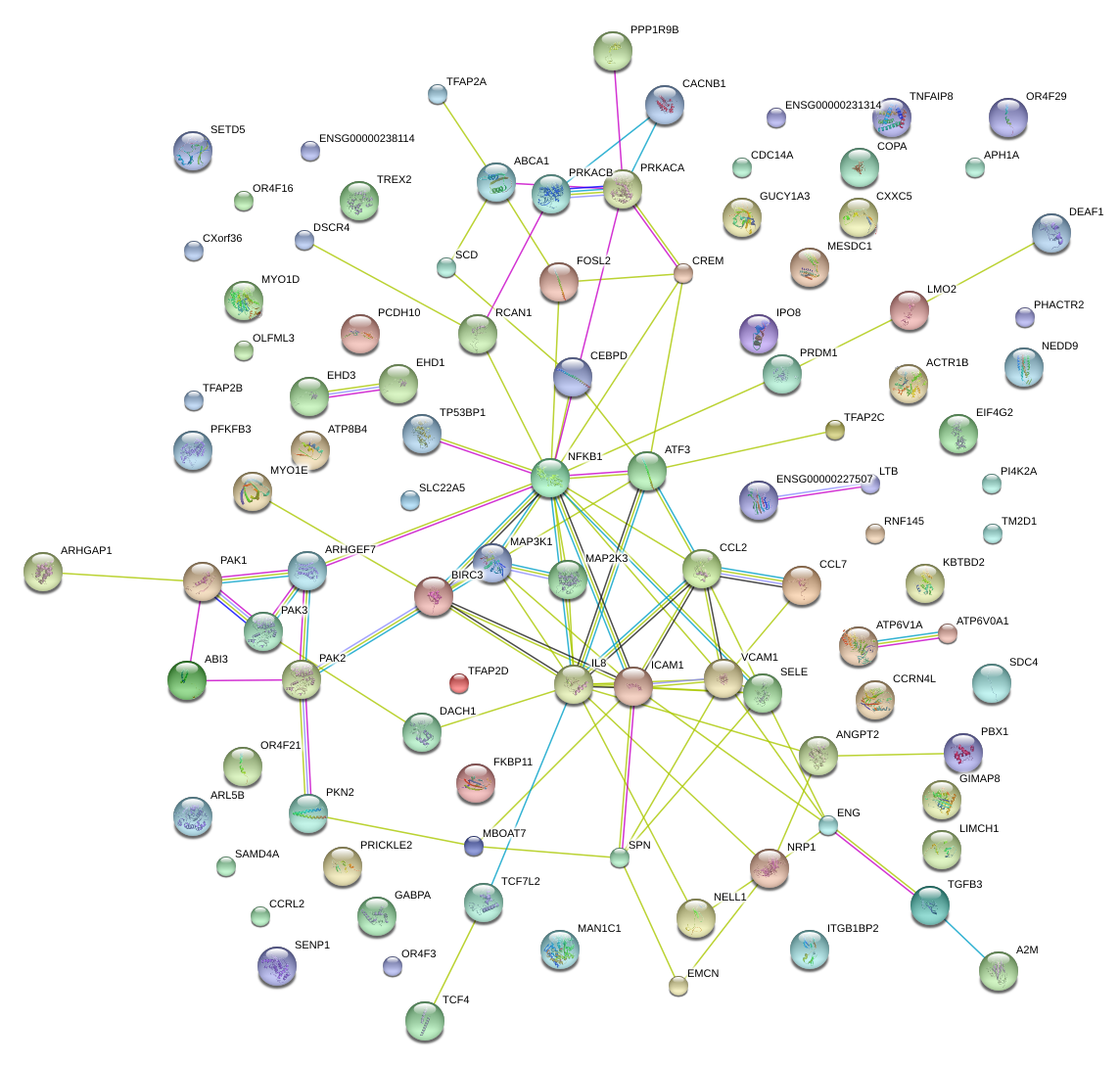
SELE
|
selectin E
|
|
chr7_+_150147882
|
2.635
|
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr19_+_10381516
|
2.634
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr1_+_101185195
|
2.612
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr6_-_11382501
|
2.337
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr10_+_102107074
|
2.301
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr11_-_10830034
|
1.995
|
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr10_+_102107256
|
1.905
|
|
|
|
|
chr10_+_102107001
|
1.839
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr3_+_46448678
|
1.776
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr1_+_114522029
|
1.718
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr2_-_26101091
|
1.631
|
|
|
|
|
chr10_+_102106958
|
1.615
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr4_+_103422822
|
1.615
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr4_+_103422730
|
1.560
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chrX_-_45060081
|
1.474
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr3_+_113465901
|
1.467
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr21_-_35899046
|
1.405
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr13_+_111855406
|
1.358
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr1_+_164528586
|
1.334
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_+_102188180
|
1.318
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr4_+_139936938
|
1.315
|
NM_012118
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae)
|
|
chr17_+_40610940
|
1.285
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr16_+_29674564
|
1.285
|
|
SPN
|
sialophorin
|
|
chr17_+_40610897
|
1.284
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr15_-_59665069
|
1.273
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr9_-_107690526
|
1.270
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr7_+_150148078
|
1.269
|
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr10_+_18948276
|
1.203
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr8_-_48650683
|
1.164
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr17_+_40610875
|
1.153
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr6_-_10412606
|
1.142
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr2_+_28616346
|
1.128
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr17_+_47288068
|
1.126
|
|
ABI3
|
ABI family, member 3
|
|
chr4_+_74606222
|
1.125
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr4_+_41540187
|
1.113
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr21_+_27107731
|
1.108
|
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr17_+_40610905
|
1.105
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr11_-_33890931
|
1.094
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr7_-_32931409
|
1.079
|
NM_015483
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr4_+_41700736
|
1.064
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_-_32931363
|
1.055
|
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr4_-_101439071
|
1.028
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr13_-_72441312
|
1.027
|
NM_004392
NM_080759
NM_080760
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr17_+_21191347
|
1.024
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr19_+_10381763
|
1.017
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr9_-_130616592
|
1.003
|
|
ENG
|
endoglin
|
|
chr21_+_27107522
|
0.989
|
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr1_+_212738675
|
0.988
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr17_+_32582295
|
0.971
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr3_-_64211111
|
0.960
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr2_+_28616360
|
0.956
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr17_+_32597239
|
0.940
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr18_-_53068755
|
0.938
|
|
TCF4
|
transcription factor 4
|
|
chr8_-_6420454
|
0.932
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr12_-_48499840
|
0.929
|
|
SENP1
|
SUMO1/sentrin specific peptidase 1
|
|
chr20_-_43977023
|
0.926
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr4_+_103422553
|
0.926
|
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
|
|
chr7_+_150147936
|
0.895
|
NM_175571
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr5_+_56111379
|
0.881
|
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr15_-_59665053
|
0.879
|
|
MYO1E
|
myosin IE
|
|
chr5_+_140734767
|
0.873
|
NM_018917
NM_032053
|
PCDHGA4
|
protocadherin gamma subfamily A, 4
|
|
chr10_+_35484829
|
0.873
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr1_-_150241409
|
0.862
|
NM_001077628
NM_001243771
NM_001243772
NM_016022
|
APH1A
|
anterior pharynx defective 1 homolog A (C. elegans)
|
|
chr19_-_54692132
|
0.858
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr10_+_99400469
|
0.847
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr3_+_113465860
|
0.838
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr5_+_118691692
|
0.832
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr15_-_43785368
|
0.826
|
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr5_+_180794268
|
0.825
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr6_-_31550065
|
0.820
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr14_-_76448091
|
0.811
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr10_+_6186878
|
0.810
|
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr10_-_33623978
|
0.809
|
|
NRP1
|
neuropilin 1
|
|
chr6_+_106534188
|
0.802
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr4_+_134070443
|
0.797
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr11_-_33891258
|
0.797
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr5_-_158635080
|
0.793
|
NM_001199381
|
RNF145
|
ring finger protein 145
|
|
chr1_+_25943958
|
0.786
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr12_-_9268440
|
0.780
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_100818052
|
0.775
|
|
CDC14A
|
CDC14 cell division cycle 14 homolog A (S. cerevisiae)
|
|
chr2_-_54198055
|
0.775
|
|
|
|
|
chr1_-_160313056
|
0.774
|
|
COPA
|
coatomer protein complex, subunit alpha
|
|
chr11_-_33891361
|
0.771
|
NM_001142316
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr12_-_9268513
|
0.755
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_-_150241210
|
0.752
|
|
APH1A
|
anterior pharynx defective 1 homolog A (C. elegans)
|
|
chr2_+_28615668
|
0.744
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr17_-_31204071
|
0.742
|
|
MYO1D
|
myosin ID
|
|
chr1_+_84609941
|
0.742
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr12_-_9268522
|
0.742
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr7_-_32931189
|
0.730
|
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr17_+_40610857
|
0.729
|
NM_001130020
NM_001130021
NM_005177
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr5_+_139060295
|
0.727
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr11_-_64647136
|
0.724
|
|
EHD1
|
EH-domain containing 1
|
|
chr3_+_196466806
|
0.715
|
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr6_+_143999058
|
0.711
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr5_-_158634641
|
0.707
|
NM_001199382
|
RNF145
|
ring finger protein 145
|
|
chr3_+_9476506
|
0.705
|
|
|
|
|
chr11_-_46717657
|
0.704
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr4_+_156587861
|
0.703
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr10_+_102106771
|
0.697
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr14_+_55034329
|
0.696
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chrX_+_70521627
|
0.694
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr21_+_27107671
|
0.694
|
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr15_+_81293286
|
0.694
|
NM_022566
|
MESDC1
|
mesoderm development candidate 1
|
|
chr3_+_9439533
|
0.678
|
|
SETD5
|
SET domain containing 5
|
|
chr15_-_50411405
|
0.676
|
NM_024837
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr12_-_30848744
|
0.662
|
|
IPO8
|
importin 8
|
|
chr1_-_62190740
|
0.662
|
|
TM2D1
|
TM2 domain containing 1
|
|
chr3_+_113465910
|
0.657
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr21_+_41117333
|
0.657
|
NM_001080444
|
IGSF5
|
immunoglobulin superfamily, member 5
|
|
chr8_-_22550708
|
0.654
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr12_-_110434028
|
0.649
|
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chr10_+_102121371
|
0.649
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr13_+_97928414
|
0.648
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr16_+_30406432
|
0.646
|
NM_001214909
|
ZNF48
|
zinc finger protein 48
|
|
chr11_-_71791734
|
0.645
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr17_-_73401775
|
0.643
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr8_+_90770201
|
0.642
|
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chr13_+_97928196
|
0.637
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_+_10353829
|
0.633
|
|
MARCH6
|
membrane-associated ring finger (C3HC4) 6
|
|
chr13_+_97874704
|
0.632
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_+_173315313
|
0.631
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr8_+_90769974
|
0.630
|
NM_003821
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chr3_-_71179736
|
0.626
|
|
FOXP1
|
forkhead box P1
|
|
chr2_+_85766286
|
0.625
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr1_-_47082529
|
0.625
|
NM_201403
|
MOB3C
|
MOB kinase activator 3C
|
|
chr8_-_117023
|
0.624
|
NM_001005504
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21
|
|
chr8_-_22550057
|
0.623
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr1_-_221910801
|
0.623
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr12_-_10251520
|
0.620
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr5_-_158634833
|
0.615
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr14_+_100531764
|
0.613
|
|
EVL
|
Enah/Vasp-like
|
|
chr6_-_47253962
|
0.613
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr9_+_116343169
|
0.613
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_116342939
|
0.610
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr6_-_47254143
|
0.608
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr22_+_45704821
|
0.608
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr1_-_21605984
|
0.606
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr4_+_74735107
|
0.604
|
NM_001511
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr17_+_57785010
|
0.603
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr14_+_53019865
|
0.602
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr8_+_86157693
|
0.600
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr10_+_30723050
|
0.599
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr3_+_100328432
|
0.597
|
NM_032787
|
GPR128
|
G protein-coupled receptor 128
|
|
chr14_+_100531745
|
0.596
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr6_-_43595079
|
0.596
|
|
GTPBP2
|
GTP binding protein 2
|
|
chr22_+_38453478
|
0.594
|
|
PICK1
|
protein interacting with PRKCA 1
|
|
chr1_-_112046742
|
0.592
|
NM_000677
NM_020683
|
ADORA3
|
adenosine A3 receptor
|
|
chr7_-_37488894
|
0.591
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr17_-_63052707
|
0.589
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chrX_-_48815582
|
0.584
|
|
OTUD5
|
OTU domain containing 5
|
|
chr4_-_185395616
|
0.584
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr9_+_120466607
|
0.582
|
|
TLR4
|
toll-like receptor 4
|
|
chr1_-_155948206
|
0.577
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr8_+_68864589
|
0.574
|
NM_024870
NM_025170
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr10_+_73555527
|
0.564
|
NM_001171933
NM_001171934
|
CDH23
|
cadherin-related 23
|
|
chr12_-_89746238
|
0.564
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr2_+_28615771
|
0.561
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr20_-_42839423
|
0.561
|
NM_016470
|
C20orf111
|
chromosome 20 open reading frame 111
|
|
chr7_-_37488408
|
0.556
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr4_-_16900256
|
0.555
|
|
LDB2
|
LIM domain binding 2
|
|
chr3_-_129035069
|
0.555
|
NM_006026
|
H1FX
|
H1 histone family, member X
|
|
chr1_-_155948317
|
0.555
|
NM_001162383
NM_001162384
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr7_-_37488554
|
0.553
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr10_-_98480174
|
0.550
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr14_-_21852161
|
0.550
|
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr5_-_39425030
|
0.550
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr12_-_9268475
|
0.550
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr14_-_21852085
|
0.549
|
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr4_-_16900348
|
0.546
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_16900028
|
0.544
|
|
LDB2
|
LIM domain binding 2
|
|
chr20_-_30311693
|
0.541
|
|
BCL2L1
|
BCL2-like 1
|
|
chr17_-_73389708
|
0.541
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr4_+_156588811
|
0.540
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr13_+_46039024
|
0.540
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr10_-_116286570
|
0.540
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr10_+_30722856
|
0.537
|
NM_001244134
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr4_+_42399855
|
0.531
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr6_-_154831788
|
0.529
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr1_-_91487543
|
0.527
|
|
ZNF644
|
zinc finger protein 644
|
|
chr6_+_14118002
|
0.526
|
|
CD83
|
CD83 molecule
|
|
chr19_+_45251933
|
0.526
|
NM_005178
|
BCL3
|
B-cell CLL/lymphoma 3
|
|
chr10_+_37414784
|
0.522
|
NM_052997
|
ANKRD30A
|
ankyrin repeat domain 30A
|
|
chr5_-_32403310
|
0.521
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr1_+_218519576
|
0.517
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr10_-_33623771
|
0.517
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr10_-_103578179
|
0.514
|
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase)
|
|
chr17_-_35656691
|
0.514
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr6_+_14117857
|
0.513
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr1_+_153175918
|
0.513
|
NM_001010857
|
LELP1
|
late cornified envelope-like proline-rich 1
|
|
chr20_-_42839295
|
0.511
|
|
C20orf111
|
chromosome 20 open reading frame 111
|
|
chr11_-_61124223
|
0.505
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chrX_+_149613719
|
0.505
|
NM_001177466
NM_005491
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr4_+_41540320
|
0.503
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_16259046
|
0.502
|
|
|
|
|
chr2_-_26101173
|
0.501
|
|
ASXL2
|
additional sex combs like 2 (Drosophila)
|
|
chr21_-_39870303
|
0.495
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr21_-_47704166
|
0.494
|
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr14_-_35344852
|
0.491
|
NM_013448
NM_182648
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|