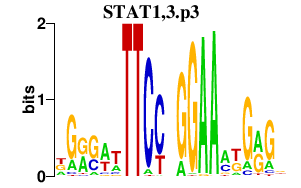
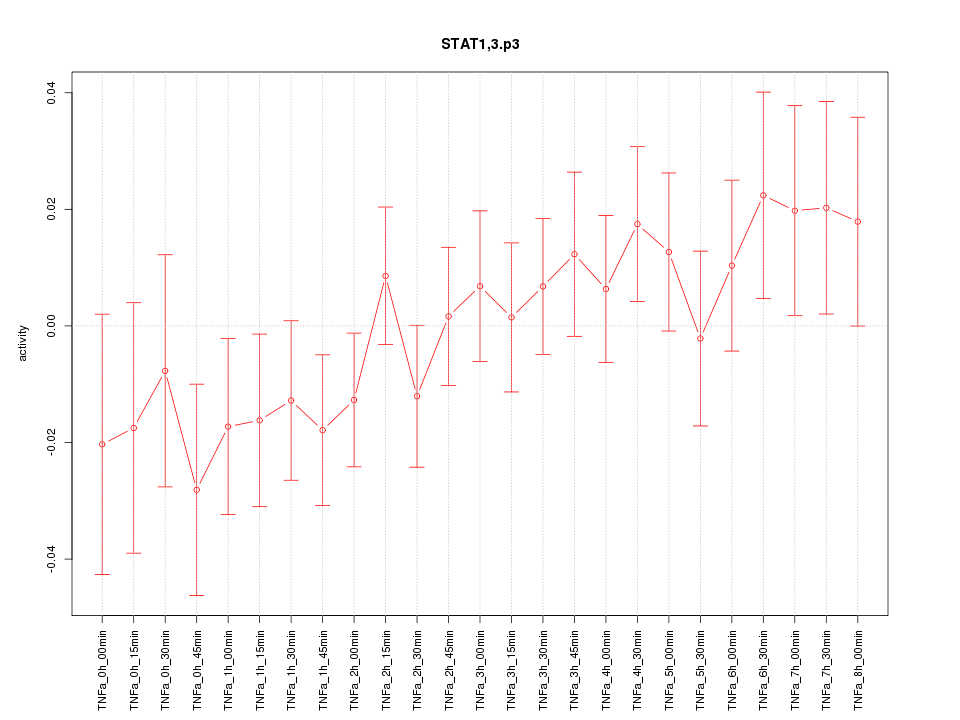
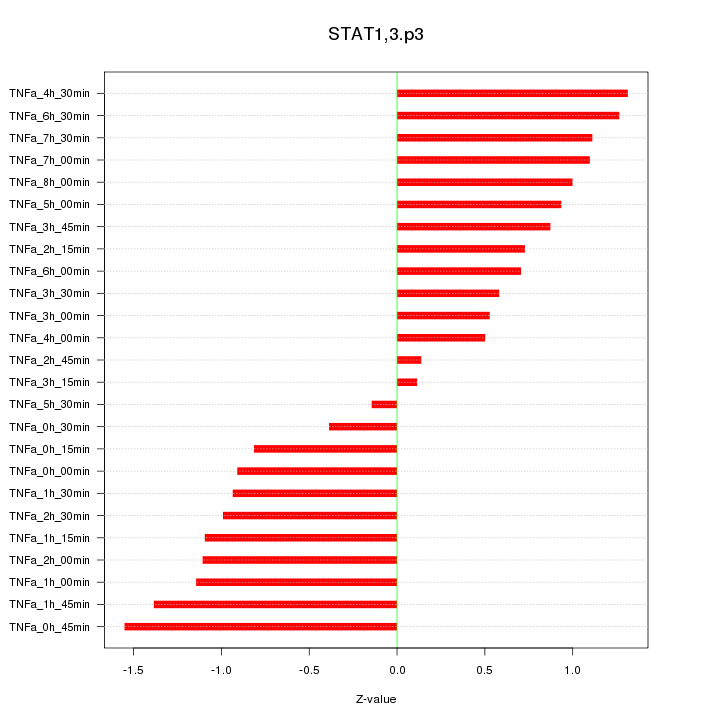
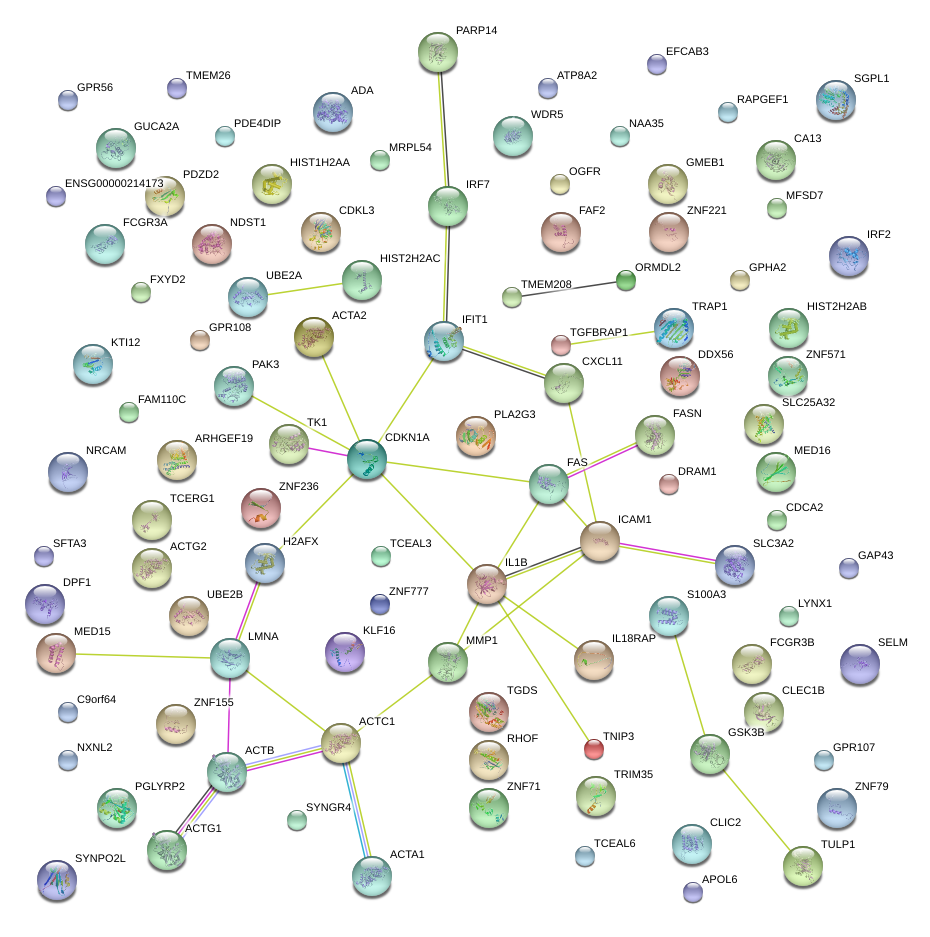
|
chr4_-_76957139
|
7.620
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr3_+_122399665
|
7.199
|
NM_017554
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr1_-_153521717
|
5.852
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr19_+_10381763
|
5.239
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr22_+_36044412
|
2.539
|
NM_030641
|
APOL6
|
apolipoprotein L, 6
|
|
chrX_-_154563735
|
2.517
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr4_-_122148620
|
2.512
|
NM_001244764
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr12_+_102271082
|
2.223
|
NM_018370
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr10_+_90751044
|
2.166
|
|
FAS
|
Fas (TNF receptor superfamily, member 6)
|
|
chr10_-_90750955
|
2.143
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr8_+_86158004
|
2.005
|
|
CA13
|
carbonic anhydrase XIII
|
|
chr8_+_86157693
|
1.988
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr11_-_102668822
|
1.904
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr22_-_31503484
|
1.876
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr22_+_20861875
|
1.820
|
|
MED15
|
mediator complex subunit 15
|
|
chr5_+_149887673
|
1.750
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr22_+_20861884
|
1.738
|
NM_001003891
NM_015889
|
MED15
|
mediator complex subunit 15
|
|
chr22_+_20861886
|
1.733
|
|
MED15
|
mediator complex subunit 15
|
|
chr10_+_91152321
|
1.629
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr22_+_20861898
|
1.562
|
|
MED15
|
mediator complex subunit 15
|
|
chr6_+_36646455
|
1.372
|
NM_000389
NM_001220778
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr1_-_144994943
|
1.275
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_+_10381516
|
1.239
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr1_-_144994839
|
1.222
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr11_+_62623466
|
1.122
|
NM_001012662
NM_001012664
NM_002394
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr13_-_95248482
|
1.085
|
NM_014305
|
TGDS
|
TDP-glucose 4,6-dehydratase
|
|
chr20_+_61436160
|
1.025
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr9_+_137004913
|
1.017
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr11_-_102668892
|
0.977
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr7_-_108096693
|
0.954
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr3_-_119582422
|
0.944
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr1_+_156084579
|
0.792
|
|
LMNA
|
lamin A/C
|
|
chr19_+_3762671
|
0.782
|
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr5_+_31799030
|
0.762
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr11_-_615920
|
0.759
|
|
IRF7
|
interferon regulatory factor 7
|
|
chr9_+_132897342
|
0.744
|
|
GPR107
|
G protein-coupled receptor 107
|
|
chr22_-_31536339
|
0.719
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr9_-_86571631
|
0.718
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr1_-_144994908
|
0.697
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr12_+_56211862
|
0.692
|
|
ORMDL2
|
ORM1-like 2 (S. cerevisiae)
|
|
chr17_+_60457913
|
0.663
|
NM_173503
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr6_+_36646489
|
0.655
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr5_-_133706717
|
0.630
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr19_+_57106622
|
0.623
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr12_+_56211718
|
0.622
|
NM_014182
|
ORMDL2
|
ORM1-like 2 (S. cerevisiae)
|
|
chr13_-_95248401
|
0.620
|
|
TGDS
|
TDP-glucose 4,6-dehydratase
|
|
chr10_-_63212924
|
0.609
|
|
TMEM26
|
transmembrane protein 26
|
|
chr2_+_103035148
|
0.603
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr16_+_67261010
|
0.595
|
NM_014187
|
TMEM208
|
transmembrane protein 208
|
|
chr12_-_10151689
|
0.585
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr9_-_134585107
|
0.576
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr5_+_175875372
|
0.574
|
|
FAF2
|
Fas associated factor family member 2
|
|
chr19_+_3762650
|
0.553
|
NM_172251
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr17_-_76183074
|
0.553
|
NM_003258
|
TK1
|
thymidine kinase 1, soluble
|
|
chr5_+_133706869
|
0.546
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr9_+_88556102
|
0.525
|
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr11_-_62623212
|
0.512
|
|
SNHG1
|
small nucleolar RNA host gene 1 (non-protein coding)
|
|
chr8_+_25316714
|
0.506
|
|
CDCA2
|
cell division cycle associated 2
|
|
chr1_-_52499432
|
0.490
|
NM_138417
|
KTI12
|
KTI12 homolog, chromatin associated (S. cerevisiae)
|
|
chr9_+_88556056
|
0.488
|
NM_024635
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr16_-_3767449
|
0.482
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr6_-_35480635
|
0.480
|
NM_003322
|
TULP1
|
tubby like protein 1
|
|
chr16_+_57653649
|
0.459
|
|
GPR56
|
G protein-coupled receptor 56
|
|
chr5_+_145826866
|
0.448
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr2_-_113594325
|
0.447
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr19_-_6737603
|
0.444
|
NM_001080452
|
GPR108
|
G protein-coupled receptor 108
|
|
chr13_+_99058147
|
0.437
|
|
|
|
|
chr10_+_72575724
|
0.437
|
|
SGPL1
|
sphingosine-1-phosphate lyase 1
|
|
chr11_-_615998
|
0.422
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr11_-_117695408
|
0.409
|
NM_001680
|
FXYD2
|
FXYD domain containing ion transport regulator 2
|
|
chr19_-_38720312
|
0.406
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr19_+_44331443
|
0.403
|
NM_181845
|
ZNF283
|
zinc finger protein 283
|
|
chr2_-_46384
|
0.393
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr8_-_104427373
|
0.392
|
NM_030780
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr4_-_683107
|
0.383
|
|
MFSD7
|
major facilitator superfamily domain containing 7
|
|
chr20_-_43280359
|
0.383
|
NM_000022
|
ADA
|
adenosine deaminase
|
|
chr19_+_48867650
|
0.379
|
NM_012451
|
SYNGR4
|
synaptogyrin 4
|
|
chr19_+_44488331
|
0.378
|
NM_003445
NM_198089
|
ZNF155
|
zinc finger protein 155
|
|
chrX_+_102862833
|
0.377
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr8_-_27168783
|
0.367
|
NM_171982
|
TRIM35
|
tripartite motif containing 35
|
|
chr3_+_115342150
|
0.362
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr11_-_118966175
|
0.360
|
NM_002105
|
H2AFX
|
H2A histone family, member X
|
|
chr11_-_615649
|
0.354
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr1_-_161519817
|
0.353
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr22_+_20861964
|
0.350
|
|
MED15
|
mediator complex subunit 15
|
|
chr5_+_175875386
|
0.349
|
|
FAF2
|
Fas associated factor family member 2
|
|
chr14_-_36982976
|
0.348
|
NM_001101341
|
SFTA3
|
surfactant associated 3
|
|
chr8_+_25316736
|
0.346
|
|
CDCA2
|
cell division cycle associated 2
|
|
chr13_+_25946084
|
0.339
|
NM_016529
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr8_-_143859177
|
0.330
|
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr12_-_122231554
|
0.330
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr10_-_75415748
|
0.330
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr4_-_185395616
|
0.330
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr1_-_16539103
|
0.329
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr9_+_130186714
|
0.327
|
|
ZNF79
|
zinc finger protein 79
|
|
chr11_-_64703334
|
0.324
|
NM_130769
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chr7_-_149157866
|
0.316
|
NM_015694
|
ZNF777
|
zinc finger protein 777
|
|
chr1_-_42630386
|
0.307
|
NM_033553
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin)
|
|
chr7_-_44613488
|
0.304
|
|
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 56
|
|
chr19_-_15590203
|
0.290
|
NM_052890
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr19_-_893215
|
0.289
|
NM_005481
|
MED16
|
mediator complex subunit 16
|
|
chr19_-_40331153
|
0.284
|
|
FBL
|
fibrillarin
|
|
chr16_-_1470709
|
0.282
|
|
C16orf91
|
chromosome 16 open reading frame 91
|
|
chr11_+_32112414
|
0.281
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr20_+_37101457
|
0.280
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr6_+_31913852
|
0.276
|
|
CFB
|
complement factor B
|
|
chr5_+_175875355
|
0.274
|
NM_014613
|
FAF2
|
Fas associated factor family member 2
|
|
chr9_+_136287119
|
0.269
|
NM_139025
NM_139026
NM_139027
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr6_+_36644236
|
0.267
|
NM_001220777
NM_078467
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr1_-_161519526
|
0.263
|
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr16_+_4674829
|
0.262
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr6_-_25874439
|
0.255
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr17_+_32646058
|
0.253
|
NM_005623
|
CCL8
|
chemokine (C-C motif) ligand 8
|
|
chr3_+_128598520
|
0.252
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr3_-_118959723
|
0.249
|
NM_003778
NM_212543
|
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4
|
|
chr8_-_134115309
|
0.247
|
NM_001045556
NM_001045557
|
SLA
|
Src-like-adaptor
|
|
chr19_-_6737565
|
0.241
|
|
GPR108
|
G protein-coupled receptor 108
|
|
chr2_+_171785780
|
0.241
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr19_+_44716683
|
0.240
|
NM_182490
|
ZNF227
|
zinc finger protein 227
|
|
chr6_+_41303527
|
0.240
|
NM_001199509
NM_001199510
NM_004828
|
NCR2
|
natural cytotoxicity triggering receptor 2
|
|
chr18_-_52626636
|
0.240
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr4_+_119809978
|
0.233
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr20_+_35973083
|
0.230
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr16_+_70207927
|
0.229
|
NM_173619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr22_+_18121540
|
0.223
|
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator)
|
|
chr2_-_11810254
|
0.222
|
NM_012344
|
NTSR2
|
neurotensin receptor 2
|
|
chr14_+_55168831
|
0.222
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr8_-_18666295
|
0.222
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr16_-_74455239
|
0.215
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr18_+_55714457
|
0.215
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr10_+_86004808
|
0.212
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr3_+_187086167
|
0.208
|
NM_022147
|
RTP4
|
receptor (chemosensory) transporter protein 4
|
|
chr9_-_131038196
|
0.207
|
NM_004486
|
GOLGA2
|
golgin A2
|
|
chr2_+_170366211
|
0.206
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr17_-_6983503
|
0.205
|
NM_006344
NM_182906
|
CLEC10A
|
C-type lectin domain family 10, member A
|
|
chr14_+_73957670
|
0.204
|
|
C14orf169
|
chromosome 14 open reading frame 169
|
|
chr1_+_54519259
|
0.201
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr19_+_44598479
|
0.201
|
NM_013398
|
ZNF224
|
zinc finger protein 224
|
|
chr3_+_130612806
|
0.199
|
NM_001199179
NM_001001485
NM_001199183
NM_001199184
NM_001199185
NM_014382
NM_001001486
NM_001001487
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr11_+_46694514
|
0.194
|
|
ATG13
|
ATG13 autophagy related 13 homolog (S. cerevisiae)
|
|
chr3_+_128598471
|
0.194
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr6_-_111136616
|
0.194
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr3_+_115342302
|
0.192
|
|
GAP43
|
growth associated protein 43
|
|
chr3_+_128598440
|
0.191
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr8_+_36641841
|
0.191
|
NM_001031836
|
KCNU1
|
potassium channel, subfamily U, member 1
|
|
chr3_+_128598459
|
0.191
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr9_+_130186652
|
0.181
|
NM_007135
|
ZNF79
|
zinc finger protein 79
|
|
chr19_-_893184
|
0.180
|
|
MED16
|
mediator complex subunit 16
|
|
chr5_+_178368193
|
0.176
|
NM_001178089
NM_001178090
NM_182594
|
ZNF454
|
zinc finger protein 454
|
|
chr20_+_2795613
|
0.175
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr20_+_24449834
|
0.174
|
NM_024893
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr6_+_31540092
|
0.174
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr11_-_118966103
|
0.173
|
|
H2AFX
|
H2A histone family, member X
|
|
chr19_-_6737589
|
0.172
|
|
GPR108
|
G protein-coupled receptor 108
|
|
chr4_-_48136245
|
0.169
|
NM_003328
|
TXK
|
TXK tyrosine kinase
|
|
chr18_+_268147
|
0.160
|
|
|
|
|
chr4_-_176733901
|
0.158
|
|
GPM6A
|
glycoprotein M6A
|
|
chr1_-_212004113
|
0.156
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr5_-_138725544
|
0.154
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr4_+_3443659
|
0.154
|
|
HGFAC
|
HGF activator
|
|
chr14_+_67707825
|
0.146
|
NM_022474
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr6_+_54711568
|
0.146
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr9_-_137809710
|
0.145
|
NM_002003
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1
|
|
chr10_+_95848811
|
0.145
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr16_-_29910340
|
0.145
|
NM_001114099
NM_001114100
NM_001243332
NM_001243333
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr8_-_143859638
|
0.145
|
NM_023946
NM_177457
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr17_+_7462064
|
0.145
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr3_+_128598332
|
0.141
|
NM_014049
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr10_-_61469322
|
0.140
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr11_+_61447816
|
0.140
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr2_+_149894980
|
0.138
|
NM_177964
|
LYPD6B
|
LY6/PLAUR domain containing 6B
|
|
chr4_+_3443691
|
0.138
|
NM_001528
|
HGFAC
|
HGF activator
|
|
chr17_-_3030844
|
0.136
|
NM_003555
|
OR1G1
|
olfactory receptor, family 1, subfamily G, member 1
|
|
chr3_+_37284680
|
0.135
|
NM_001172713
NM_002078
|
GOLGA4
|
golgin A4
|
|
chr17_-_39254374
|
0.134
|
NM_031960
|
KRTAP4-8
|
keratin associated protein 4-8
|
|
chr5_+_140261792
|
0.131
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr5_+_33441046
|
0.130
|
|
TARS
|
threonyl-tRNA synthetase
|
|
chr20_-_34243118
|
0.125
|
|
RBM12
|
RNA binding motif protein 12
|
|
chr14_-_68141328
|
0.124
|
NM_006370
|
VTI1B
|
vesicle transport through interaction with t-SNAREs homolog 1B (yeast)
|
|
chr11_-_129062092
|
0.123
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr20_-_6034617
|
0.120
|
NM_152611
|
LRRN4
|
leucine rich repeat neuronal 4
|
|
chr4_-_21546278
|
0.118
|
NM_001035004
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr1_+_46153846
|
0.112
|
NM_016486
|
TMEM69
|
transmembrane protein 69
|
|
chr9_-_23821477
|
0.111
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr10_+_71029755
|
0.110
|
NM_033497
NM_033498
NM_033500
|
HK1
|
hexokinase 1
|
|
chr6_+_31913763
|
0.109
|
|
CFB
|
complement factor B
|
|
chr9_+_74764394
|
0.109
|
|
GDA
|
guanine deaminase
|
|
chr22_-_24096551
|
0.107
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr19_+_42300521
|
0.107
|
NM_001815
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3
|
|
chr6_+_53883713
|
0.105
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr12_-_75603214
|
0.104
|
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr19_+_10828813
|
0.102
|
|
DNM2
|
dynamin 2
|
|
chr9_+_74764382
|
0.100
|
|
GDA
|
guanine deaminase
|
|
chr1_+_50569581
|
0.099
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr19_+_49977465
|
0.098
|
NM_001204502
NM_001459
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr1_+_156123321
|
0.098
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr2_+_30670082
|
0.098
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr1_+_34632483
|
0.092
|
NM_032884
|
C1orf94
|
chromosome 1 open reading frame 94
|
|
chr19_-_14049167
|
0.091
|
NM_024825
|
PODNL1
|
podocan-like 1
|
|
chr3_+_128598497
|
0.090
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|