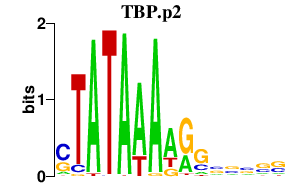
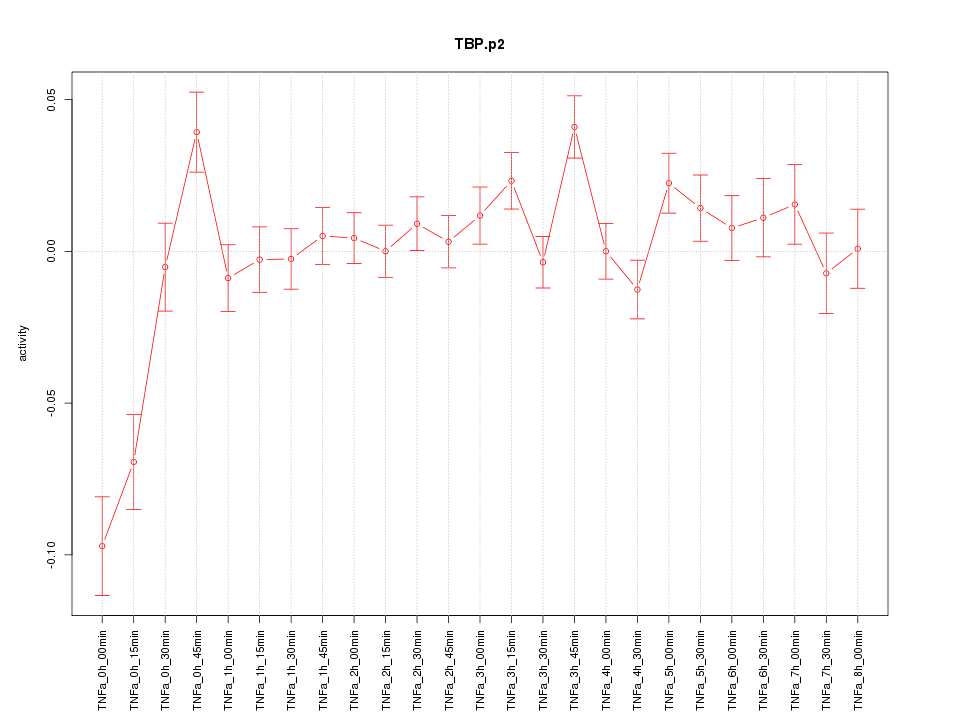
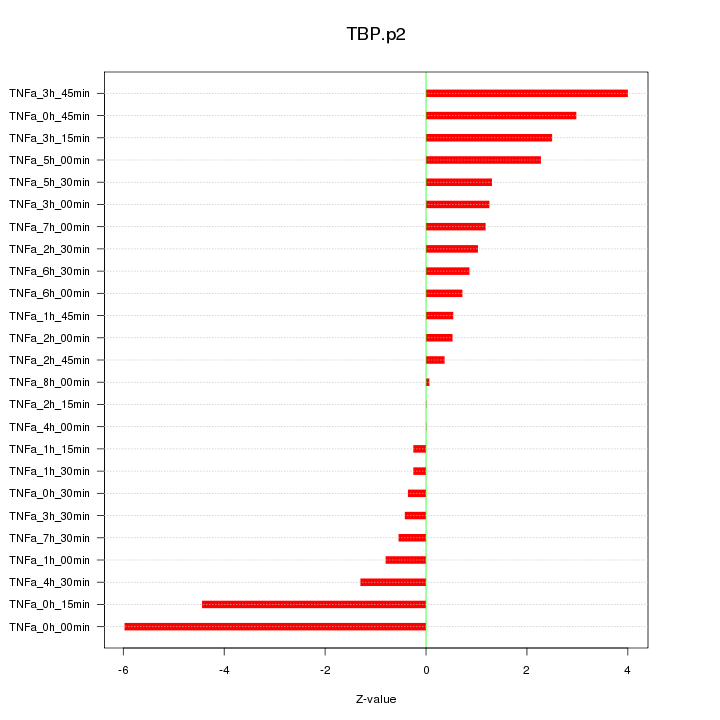
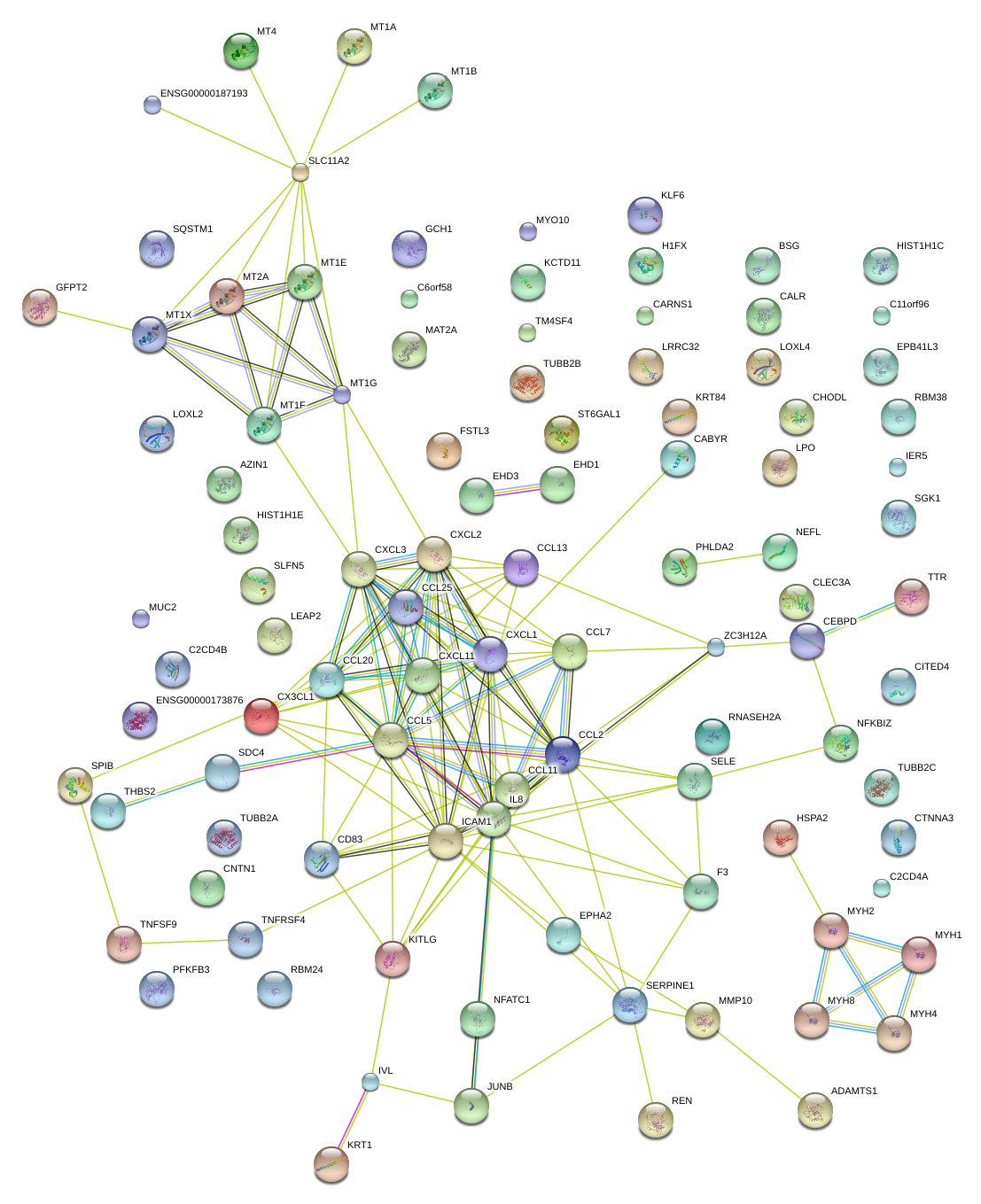
|
chr1_-_169703178
|
20.475
|
NM_000450
|
SELE
|
selectin E
|
|
chr19_+_10381763
|
19.383
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr21_+_26934440
|
13.068
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr11_-_102651321
|
10.707
|
NM_002425
|
MMP10
|
matrix metallopeptidase 10 (stromelysin 2)
|
|
chr15_+_62359175
|
10.279
|
NM_207322
|
C2CD4A
|
C2 calcium-dependent domain containing 4A
|
|
chr1_-_95007139
|
9.463
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr4_-_74904329
|
9.346
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr4_+_74606222
|
8.145
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr15_-_62457481
|
7.928
|
NM_001007595
|
C2CD4B
|
C2 calcium-dependent domain containing 4B
|
|
chr16_+_57406398
|
7.696
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr4_+_74735107
|
7.641
|
NM_001511
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr2_+_228678556
|
7.396
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr1_+_37940180
|
6.246
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr4_-_74964836
|
5.547
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr6_+_14118002
|
5.165
|
|
CD83
|
CD83 molecule
|
|
chr14_-_55369163
|
5.038
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr17_+_32582295
|
4.934
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr6_+_127898318
|
4.523
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr19_+_676346
|
4.451
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr19_+_12902295
|
4.303
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr11_-_64646053
|
4.141
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr19_+_6531009
|
3.910
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr5_-_179780314
|
3.889
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr1_-_41328019
|
3.850
|
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr8_-_48650703
|
3.791
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr8_-_23261632
|
3.759
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr19_+_8117645
|
3.745
|
NM_001201359
NM_005624
|
CCL25
|
chemokine (C-C motif) ligand 25
|
|
chr10_+_6244839
|
3.651
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr1_+_181057835
|
3.580
|
|
IER5
|
immediate early response 5
|
|
chr7_+_100770378
|
3.570
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr17_+_32612686
|
3.384
|
NM_002986
|
CCL11
|
chemokine (C-C motif) ligand 11
|
|
chr16_+_56685810
|
3.348
|
NM_005947
|
MT1B
|
metallothionein 1B
|
|
chr11_+_43963809
|
3.334
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr5_+_179247841
|
3.291
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr19_+_10381516
|
3.243
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr20_+_55966440
|
3.216
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr17_+_32597239
|
3.098
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr18_+_21718930
|
3.098
|
NM_012189
NM_138643
NM_153768
NM_153769
NM_153770
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr20_-_43977023
|
3.074
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr1_+_37940129
|
3.007
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr3_+_101546833
|
2.957
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr5_+_132209354
|
2.950
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr5_+_179247903
|
2.861
|
|
SQSTM1
|
sequestosome 1
|
|
chr18_+_21719025
|
2.840
|
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr16_+_56716378
|
2.836
|
NM_005952
|
MT1X
|
metallothionein 1X
|
|
chr11_-_76381787
|
2.835
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr17_+_33570079
|
2.742
|
NM_144975
|
SLFN5
|
schlafen family member 5
|
|
chr19_+_572568
|
2.735
|
|
BSG
|
basigin (Ok blood group)
|
|
chr19_+_572403
|
2.714
|
NM_001728
NM_198589
|
BSG
|
basigin (Ok blood group)
|
|
chr19_+_572575
|
2.690
|
|
BSG
|
basigin (Ok blood group)
|
|
chr1_-_41328002
|
2.598
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr12_-_88974094
|
2.592
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr8_-_23261626
|
2.588
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr21_-_28217692
|
2.582
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr16_+_56672577
|
2.515
|
NM_005946
|
MT1A
|
metallothionein 1A
|
|
chr5_+_179248089
|
2.468
|
|
SQSTM1
|
sequestosome 1
|
|
chr16_+_56598960
|
2.408
|
NM_032935
|
MT4
|
metallothionein 4
|
|
chr5_+_179247911
|
2.406
|
|
SQSTM1
|
sequestosome 1
|
|
chr6_-_134498973
|
2.403
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_-_2950593
|
2.367
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr12_-_52779362
|
2.320
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr1_-_1149495
|
2.315
|
NM_003327
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4
|
|
chr18_+_29171729
|
2.296
|
NM_000371
|
TTR
|
transthyretin
|
|
chr17_+_32683470
|
2.287
|
NM_005408
|
CCL13
|
chemokine (C-C motif) ligand 13
|
|
chr8_-_23261591
|
2.257
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr3_-_129035069
|
2.135
|
NM_006026
|
H1FX
|
H1 histone family, member X
|
|
chr10_-_100027943
|
2.100
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chr11_+_67183144
|
2.082
|
NM_001166222
NM_020811
|
CARNS1
|
carnosine synthase 1
|
|
chr2_+_85766286
|
2.073
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr21_-_28217272
|
2.046
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr11_-_64645925
|
1.990
|
|
EHD1
|
EH-domain containing 1
|
|
chr19_+_13049440
|
1.981
|
|
CALR
|
calreticulin
|
|
chr6_-_3227776
|
1.971
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B class IIb
|
|
chr16_+_56659679
|
1.961
|
|
MT1E
|
metallothionein 1E
|
|
chr16_+_56659584
|
1.923
|
NM_175617
|
MT1E
|
metallothionein 1E
|
|
chr5_-_16742336
|
1.880
|
|
MYO10
|
myosin X
|
|
chr17_-_34207308
|
1.860
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr1_+_37940074
|
1.805
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr8_-_24814028
|
1.719
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr6_-_169653922
|
1.703
|
|
THBS2
|
thrombospondin 2
|
|
chr8_-_48650683
|
1.701
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr3_+_149192479
|
1.670
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr10_-_69455926
|
1.664
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr1_-_16482426
|
1.655
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr8_-_103876390
|
1.652
|
NM_015878
NM_148174
|
AZIN1
|
antizyme inhibitor 1
|
|
chr19_+_13049413
|
1.651
|
NM_004343
|
CALR
|
calreticulin
|
|
chr6_-_3157696
|
1.641
|
NM_001069
|
TUBB2A
|
tubulin, beta 2A class IIa
|
|
chr4_-_76957139
|
1.622
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr18_+_77155927
|
1.598
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr19_+_13049428
|
1.584
|
|
CALR
|
calreticulin
|
|
chr3_+_186739643
|
1.582
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr14_+_65007418
|
1.575
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr19_+_50922194
|
1.549
|
NM_001243998
NM_001243999
NM_001244000
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr16_+_78056411
|
1.472
|
NM_005752
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr6_+_26156552
|
1.439
|
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr1_-_204135420
|
1.418
|
NM_000537
|
REN
|
renin
|
|
chr11_+_1074869
|
1.412
|
NM_002457
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming
|
|
chr16_-_56701904
|
1.400
|
NM_005950
|
MT1G
|
metallothionein 1G
|
|
chr12_-_53074172
|
1.357
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr1_+_152881022
|
1.349
|
NM_005547
|
IVL
|
involucrin
|
|
chr12_-_51422012
|
1.348
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr21_+_19617145
|
1.339
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr17_-_10325266
|
1.335
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr17_+_56315786
|
1.333
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr10_-_3827418
|
1.331
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr17_-_10276321
|
1.329
|
NM_003802
|
MYH13
|
myosin, heavy chain 13, skeletal muscle
|
|
chr6_+_64282449
|
1.322
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr7_+_72742154
|
1.292
|
NM_001135211
NM_003602
|
FKBP6
|
FK506 binding protein 6, 36kDa
|
|
chr11_-_615649
|
1.286
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr8_-_23261675
|
1.285
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr3_+_42727010
|
1.248
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr1_+_181057637
|
1.239
|
NM_016545
|
IER5
|
immediate early response 5
|
|
chr21_-_32201926
|
1.231
|
NM_181606
|
KRTAP7-1
|
keratin associated protein 7-1 (gene/pseudogene)
|
|
chr10_-_3827205
|
1.230
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr1_+_42619091
|
1.225
|
NM_007102
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin)
|
|
chr1_+_12916940
|
1.188
|
NM_023014
|
PRAMEF2
|
PRAME family member 2
|
|
chr1_-_206945725
|
1.185
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr17_-_79792923
|
1.181
|
NM_001007533
|
PPP1R27
|
protein phosphatase 1, regulatory subunit 27
|
|
chr8_-_23540401
|
1.181
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr1_-_201465700
|
1.178
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr6_+_36562135
|
1.164
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr12_+_52627007
|
1.151
|
|
KRT7
|
keratin 7
|
|
chr10_+_91087601
|
1.133
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr9_-_117568292
|
1.122
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr11_-_615998
|
1.121
|
NM_001572
NM_004029
|
IRF7
|
interferon regulatory factor 7
|
|
chr7_+_94285620
|
1.117
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr6_-_33771756
|
1.106
|
NM_001040109
NM_001184698
NM_002418
|
MLN
|
motilin
|
|
chr17_-_47045953
|
1.104
|
NM_004123
|
GIP
|
gastric inhibitory polypeptide
|
|
chr17_-_33700619
|
1.102
|
NM_001104587
NM_001104588
NM_001104589
NM_001104590
NM_152270
|
SLFN11
|
schlafen family member 11
|
|
chr17_-_38911524
|
1.098
|
NM_181534
|
KRT25
|
keratin 25
|
|
chr18_+_56887389
|
1.092
|
NM_001012512
NM_001012513
NM_002091
|
GRP
|
gastrin-releasing peptide
|
|
chr16_+_89989686
|
1.087
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr19_+_7030593
|
1.080
|
NM_001136507
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5
|
|
chr17_+_56270085
|
1.079
|
NM_000502
|
EPX
|
eosinophil peroxidase
|
|
chr20_-_44540685
|
1.076
|
|
PLTP
|
phospholipid transfer protein
|
|
chr6_+_26156558
|
1.062
|
NM_005321
|
HIST1H1E
|
histone cluster 1, H1e
|
|
chr12_-_11463353
|
1.062
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr2_+_85766291
|
1.041
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr7_+_94285676
|
1.028
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr17_-_79792867
|
1.028
|
|
PPP1R27
|
protein phosphatase 1, regulatory subunit 27
|
|
chr6_+_64281910
|
0.999
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr14_-_53417808
|
0.979
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr12_-_11548485
|
0.975
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr10_+_118380451
|
0.970
|
NM_005396
|
PNLIPRP2
|
pancreatic lipase-related protein 2
|
|
chr10_-_95170
|
0.953
|
NM_177987
|
TUBB8
|
tubulin, beta 8 class VIII
|
|
chr19_-_36247917
|
0.948
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr1_+_12851545
|
0.947
|
NM_023013
|
PRAMEF1
|
PRAME family member 1
|
|
chr5_+_71014989
|
0.944
|
NM_004291
|
CARTPT
|
CART prepropeptide
|
|
chr17_-_39507055
|
0.929
|
NM_004138
|
KRT33A
|
keratin 33A
|
|
chr1_-_35658430
|
0.924
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr16_+_56691854
|
0.915
|
NM_005949
|
MT1F
|
metallothionein 1F
|
|
chr11_+_65190272
|
0.911
|
|
|
|
|
chr1_-_153044083
|
0.910
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr18_+_21719447
|
0.909
|
NM_138644
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr6_-_27806116
|
0.905
|
NM_003510
|
HIST1H2AK
|
histone cluster 1, H2ak
|
|
chr3_-_195310770
|
0.898
|
|
APOD
|
apolipoprotein D
|
|
chr1_-_35658742
|
0.890
|
NM_005066
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr1_-_153518265
|
0.888
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr9_-_117880411
|
0.882
|
NM_002160
|
TNC
|
tenascin C
|
|
chr1_-_152297678
|
0.879
|
NM_002016
|
FLG
|
filaggrin
|
|
chr11_+_60970983
|
0.869
|
NM_001079807
|
PGA3
PGA4
|
pepsinogen 3, group I (pepsinogen A)
pepsinogen 4, group I (pepsinogen A)
|
|
chr13_-_114107838
|
0.866
|
NM_138430
|
ADPRHL1
|
ADP-ribosylhydrolase like 1
|
|
chr11_-_615920
|
0.861
|
|
IRF7
|
interferon regulatory factor 7
|
|
chr19_-_10514154
|
0.854
|
|
CDC37
|
cell division cycle 37 homolog (S. cerevisiae)
|
|
chr1_-_27481400
|
0.845
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr4_-_156787377
|
0.845
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr17_+_39261640
|
0.836
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr7_+_94285736
|
0.835
|
|
PEG10
|
paternally expressed 10
|
|
chr9_+_137772653
|
0.817
|
NM_004108
NM_015837
|
FCN2
|
ficolin (collagen/fibrinogen domain containing lectin) 2 (hucolin)
|
|
chr1_-_35658731
|
0.815
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr11_+_60989820
|
0.812
|
NM_001079808
|
PGA3
PGA4
|
pepsinogen 3, group I (pepsinogen A)
pepsinogen 4, group I (pepsinogen A)
|
|
chr7_-_134143804
|
0.811
|
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase)
|
|
chrX_-_139587224
|
0.808
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr17_-_39165343
|
0.800
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr15_+_76629101
|
0.782
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr2_-_75426613
|
0.773
|
NM_001058
NM_015727
|
TACR1
|
tachykinin receptor 1
|
|
chr22_-_27013959
|
0.770
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr19_+_19322763
|
0.767
|
NM_004386
|
NCAN
|
neurocan
|
|
chr12_-_52886971
|
0.766
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr17_-_39306053
|
0.764
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr8_+_22960326
|
0.761
|
NM_003841
|
TNFRSF10C
|
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain
|
|
chr5_+_145718586
|
0.761
|
NM_002700
|
POU4F3
|
POU class 4 homeobox 3
|
|
chr2_-_99917638
|
0.754
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr15_+_45406518
|
0.746
|
NM_207581
|
DUOXA2
|
dual oxidase maturation factor 2
|
|
chr19_-_7040183
|
0.744
|
NM_001164419
|
MBD3L4
|
methyl-CpG binding domain protein 3-like 4
|
|
chr12_-_52761254
|
0.740
|
NM_002283
|
KRT85
|
keratin 85
|
|
chr6_-_26017960
|
0.739
|
NM_005325
|
HIST1H1A
|
histone cluster 1, H1a
|
|
chr19_+_41594367
|
0.735
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr17_-_74733241
|
0.731
|
|
SRSF2
|
serine/arginine-rich splicing factor 2
|
|
chr11_+_68452007
|
0.729
|
|
GAL
|
galanin prepropeptide
|
|
chr1_+_153330329
|
0.726
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr3_-_99594915
|
0.726
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr6_+_111195986
|
0.724
|
NM_001634
NM_001033059
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr17_-_39341126
|
0.713
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr12_-_58165877
|
0.710
|
NM_005371
NM_023033
|
METTL1
|
methyltransferase like 1
|
|
chr8_-_23021447
|
0.706
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr16_+_56966025
|
0.706
|
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr19_-_7058644
|
0.705
|
NM_001164425
|
MBD3L3
|
methyl-CpG binding domain protein 3-like 3
|
|
chr17_+_7554239
|
0.705
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr1_+_6105980
|
0.703
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|