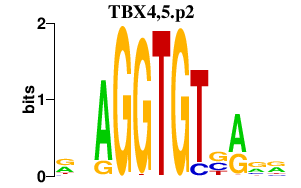
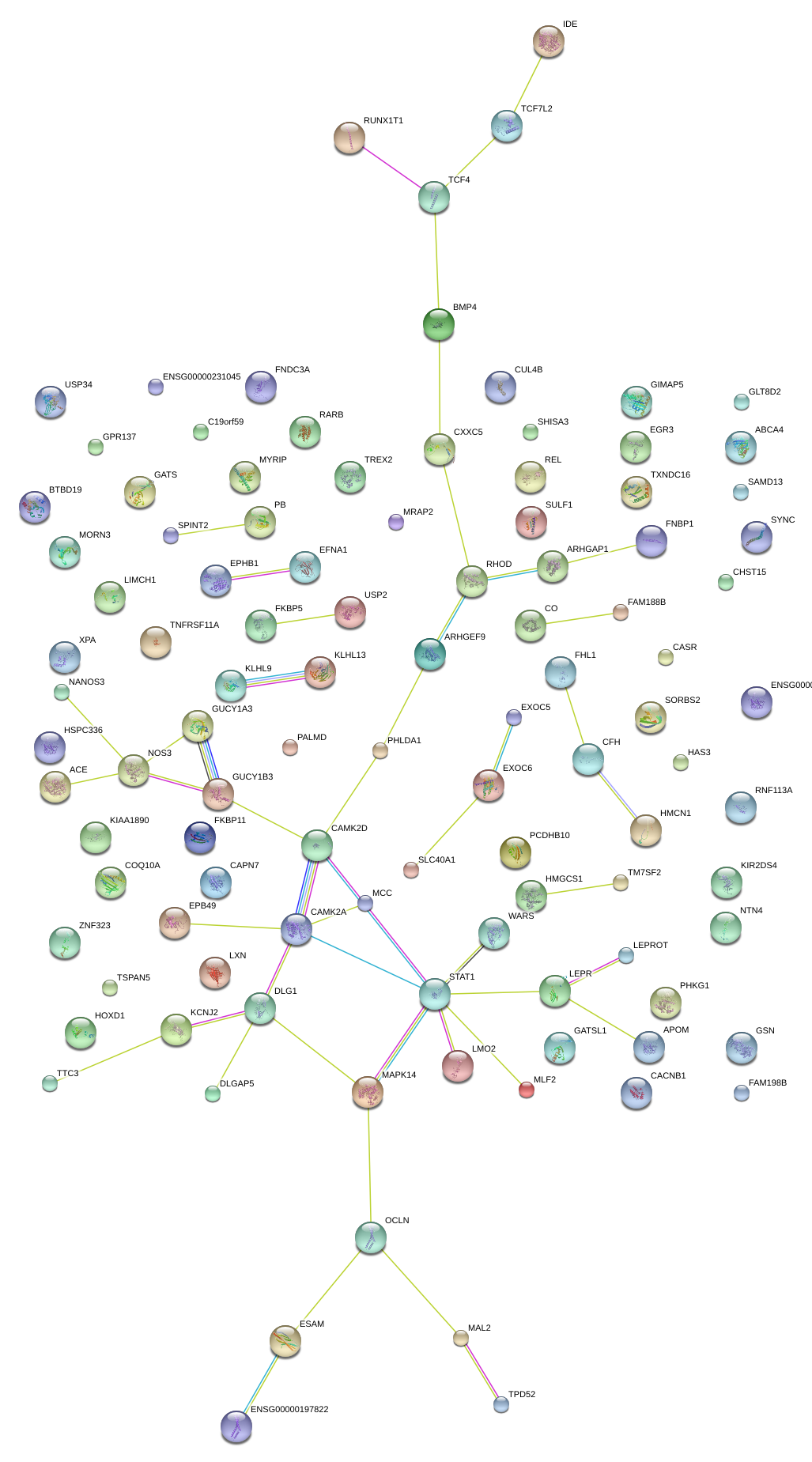
|
chr14_-_54421269
|
2.842
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr8_+_120220554
|
2.621
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr7_+_30951414
|
2.547
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr11_-_33913835
|
2.385
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chrX_-_117250624
|
1.940
|
NM_001168302
NM_001168303
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr2_-_61697846
|
1.886
|
NM_014709
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr5_-_43313492
|
1.877
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr17_+_68165660
|
1.820
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr3_+_39851028
|
1.747
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr11_-_119234628
|
1.721
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr4_+_156587861
|
1.677
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr11_-_33891258
|
1.661
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr4_-_99579540
|
1.661
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr1_+_100111430
|
1.606
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr5_-_43313581
|
1.554
|
NM_001098272
NM_002130
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr17_+_61562177
|
1.547
|
NM_001178057
NM_152830
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr2_-_61697899
|
1.546
|
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr4_+_156588811
|
1.501
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr3_-_158390266
|
1.483
|
NM_020169
|
LXN
|
latexin
|
|
chr2_+_61108702
|
1.482
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr4_+_41361621
|
1.482
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_-_132805256
|
1.421
|
|
FNBP1
|
formin binding protein 1
|
|
chr19_+_38755238
|
1.417
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr2_+_61108724
|
1.389
|
NM_002908
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr4_+_156680603
|
1.376
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr8_-_22550057
|
1.354
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr8_-_80993009
|
1.324
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr10_-_94333721
|
1.319
|
|
IDE
|
insulin-degrading enzyme
|
|
chr2_-_191878908
|
1.318
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr8_+_21912327
|
1.309
|
NM_001114136
NM_001114139
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr11_-_33891485
|
1.253
|
NM_001142315
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr21_+_38445562
|
1.253
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr19_+_38755097
|
1.242
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_-_33168356
|
1.232
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr4_-_186733372
|
1.229
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_68788118
|
1.225
|
NM_002538
|
OCLN
|
occludin
|
|
chr2_-_191878924
|
1.225
|
NM_007315
NM_139266
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr1_+_45274153
|
1.218
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr10_+_94608224
|
1.215
|
NM_019053
|
EXOC6
|
exocyst complex component 6
|
|
chr5_+_139027955
|
1.190
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr1_+_185703470
|
1.178
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr1_+_65991357
|
1.177
|
NM_001198687
NM_001198688
NM_001198689
|
LEPR
|
leptin receptor
|
|
chr18_+_59992519
|
1.169
|
NM_003839
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator
|
|
chr9_-_132805456
|
1.162
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr4_-_114682836
|
1.160
|
NM_001221
NM_172114
NM_172115
NM_172127
NM_172128
NM_172129
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr10_-_125853102
|
1.159
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr2_-_191878882
|
1.158
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr10_-_94333828
|
1.132
|
|
IDE
|
insulin-degrading enzyme
|
|
chr8_-_22549901
|
1.112
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_93029864
|
1.105
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_134514044
|
1.103
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr2_-_190445466
|
1.096
|
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr14_-_100841656
|
1.072
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr9_-_100459556
|
1.057
|
|
XPA
|
xeroderma pigmentosum, complementation group A
|
|
chrX_-_63005212
|
1.053
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr12_-_96184510
|
1.049
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr6_-_28303910
|
1.045
|
NM_001135216
NM_001243243
|
ZNF323
|
zinc finger protein 323
|
|
chr2_-_190445612
|
1.042
|
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr13_+_49550949
|
1.032
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chrX_+_135251454
|
1.025
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr5_-_112824526
|
1.017
|
NM_001085377
|
MCC
|
mutated in colorectal cancers
|
|
chr2_-_190445521
|
1.016
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr11_-_124632050
|
1.011
|
NM_138961
|
ESAM
|
endothelial cell adhesion molecule
|
|
chr4_-_159092509
|
1.005
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr19_+_7741934
|
0.995
|
NM_174918
|
C19orf59
|
chromosome 19 open reading frame 59
|
|
chr12_+_7169192
|
0.982
|
|
|
|
|
chr14_-_57735527
|
0.978
|
|
EXOC5
|
exocyst complex component 5
|
|
chr9_+_124048869
|
0.964
|
|
GSN
|
gelsolin
|
|
chr1_+_155100304
|
0.963
|
NM_004428
NM_182685
|
EFNA1
|
ephrin-A1
|
|
chr16_+_69139466
|
0.958
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr11_+_66824308
|
0.957
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr11_+_64879294
|
0.946
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr2_+_61108775
|
0.933
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr7_-_74867340
|
0.908
|
NM_001145064
|
GTF2IP1
GATSL2
|
general transcription factor IIi, pseudogene 1
GATS protein-like 2
|
|
chr1_+_65991390
|
0.905
|
|
LEPR
|
leptin receptor
|
|
chr6_+_35995583
|
0.894
|
|
MAPK14
|
mitogen-activated protein kinase 14
|
|
chr12_+_56661026
|
0.888
|
NM_001099337
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr5_+_140571903
|
0.884
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr3_-_197024879
|
0.879
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr4_-_114682594
|
0.878
|
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr12_-_104443914
|
0.878
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr3_+_25469833
|
0.871
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr8_+_70378858
|
0.862
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr18_-_52969714
|
0.857
|
NM_001243236
|
TCF4
|
transcription factor 4
|
|
chr2_+_177053306
|
0.847
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr12_-_76424474
|
0.838
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr5_+_139028483
|
0.838
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr7_+_150434679
|
0.837
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr19_+_39155209
|
0.832
|
|
|
|
|
chr7_+_150688143
|
0.831
|
NM_000603
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr1_+_84767288
|
0.826
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr5_+_139027907
|
0.824
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr7_+_150434435
|
0.822
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr4_+_42399855
|
0.811
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr1_-_94586678
|
0.808
|
NM_000350
|
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr6_+_35995585
|
0.807
|
|
MAPK14
|
mitogen-activated protein kinase 14
|
|
chr6_-_13295817
|
0.801
|
NM_001242698
|
LOC100130357
|
uncharacterized LOC100130357
|
|
chrX_-_119709618
|
0.795
|
NM_003588
|
CUL4B
|
cullin 4B
|
|
chr3_+_15247743
|
0.789
|
|
CAPN7
|
calpain 7
|
|
chr6_+_84743250
|
0.781
|
NM_138409
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr11_-_46722139
|
0.778
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr6_-_35696359
|
0.775
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr6_+_31623668
|
0.771
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr5_-_78282356
|
0.768
|
NM_000046
|
ARSB
|
arylsulfatase B
|
|
chr15_-_73076025
|
0.755
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr8_-_124553253
|
0.750
|
|
FBXO32
|
F-box protein 32
|
|
chr16_-_2581358
|
0.747
|
NM_001048212
|
CEMP1
|
cementum protein 1
|
|
chr2_+_219745243
|
0.744
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr12_-_9913415
|
0.740
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr12_+_79258794
|
0.737
|
|
SYT1
|
synaptotagmin I
|
|
chr5_-_177423242
|
0.722
|
NM_006261
|
PROP1
|
PROP paired-like homeobox 1
|
|
chr17_-_57184116
|
0.718
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr5_-_44388666
|
0.716
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr4_+_120133729
|
0.715
|
|
|
|
|
chr17_-_61523544
|
0.708
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr18_+_11981421
|
0.707
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr11_+_44785975
|
0.707
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr8_-_81083660
|
0.700
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr9_-_75567915
|
0.694
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr7_-_94285372
|
0.694
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr11_+_66824285
|
0.689
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr6_-_16761600
|
0.689
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr2_-_188419049
|
0.687
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr9_+_124048396
|
0.686
|
|
GSN
|
gelsolin
|
|
chrX_+_149531439
|
0.680
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr10_-_81205091
|
0.679
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr4_+_90816002
|
0.679
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr22_+_50528307
|
0.673
|
NM_001164104
NM_018995
|
MOV10L1
|
Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse)
|
|
chr8_-_124553448
|
0.672
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr3_+_151985828
|
0.671
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr19_+_38755422
|
0.667
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr12_+_104324214
|
0.666
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr6_-_167571315
|
0.665
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr4_-_42658883
|
0.664
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr12_+_110011499
|
0.661
|
NM_000431
NM_001114185
|
MVK
|
mevalonate kinase
|
|
chr14_-_51135041
|
0.660
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr5_+_68788387
|
0.657
|
NM_001205254
|
OCLN
|
occludin
|
|
chr2_+_207139413
|
0.651
|
NM_020923
|
ZDBF2
|
zinc finger, DBF-type containing 2
|
|
chr2_+_170590454
|
0.649
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr5_-_148930944
|
0.649
|
|
CSNK1A1
|
casein kinase 1, alpha 1
|
|
chr3_-_187454247
|
0.644
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr1_-_161102367
|
0.643
|
|
DEDD
|
death effector domain containing
|
|
chr3_+_134514300
|
0.641
|
|
EPHB1
|
EPH receptor B1
|
|
chr18_+_77659446
|
0.637
|
|
|
|
|
chr12_-_104443698
|
0.633
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr14_-_51135021
|
0.633
|
NM_021818
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr22_+_38201113
|
0.633
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr9_-_74979829
|
0.632
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr15_-_69113213
|
0.632
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr15_-_93631751
|
0.632
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr20_+_33759773
|
0.629
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr4_+_120133764
|
0.624
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr15_+_83776323
|
0.622
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr2_+_208394823
|
0.622
|
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chr12_+_104324192
|
0.620
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr6_+_35995513
|
0.619
|
|
MAPK14
|
mitogen-activated protein kinase 14
|
|
chr12_+_52416615
|
0.610
|
NM_001202233
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr19_+_48972464
|
0.610
|
NM_004228
NM_017457
|
CYTH2
|
cytohesin 2
|
|
chr1_+_38022514
|
0.606
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr7_+_104654494
|
0.606
|
NM_018682
NM_182931
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr7_-_94285426
|
0.604
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr14_-_51134862
|
0.594
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr5_+_139028025
|
0.593
|
NM_016463
|
CXXC5
|
CXXC finger protein 5
|
|
chr9_-_75568232
|
0.590
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr1_-_161102210
|
0.589
|
NM_001039712
|
DEDD
|
death effector domain containing
|
|
chr11_+_63448868
|
0.588
|
NM_006054
NM_201428
NM_201429
NM_201430
|
RTN3
|
reticulon 3
|
|
chr11_-_2170792
|
0.588
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr12_-_25055228
|
0.581
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr8_+_22436253
|
0.581
|
NM_021630
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr12_+_104324157
|
0.579
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr12_+_79257772
|
0.578
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr20_+_42574535
|
0.577
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr22_+_40441371
|
0.577
|
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr7_+_75508265
|
0.574
|
NM_001040456
NM_001040457
|
RHBDD2
|
rhomboid domain containing 2
|
|
chr20_-_3149070
|
0.565
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr17_-_27229922
|
0.565
|
NM_144683
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13
|
|
chr4_+_108745718
|
0.564
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr5_-_179718963
|
0.564
|
|
MAPK9
|
mitogen-activated protein kinase 9
|
|
chr19_+_14491920
|
0.564
|
NM_001025160
NM_001784
NM_078481
|
CD97
|
CD97 molecule
|
|
chr22_-_29075708
|
0.563
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr10_+_97515408
|
0.563
|
NM_001164178
NM_001164181
NM_001164179
NM_001164182
NM_001164183
NM_001776
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr17_-_34195745
|
0.563
|
NM_152781
|
C17orf66
|
chromosome 17 open reading frame 66
|
|
chr5_+_153418518
|
0.563
|
NM_001135037
NM_001242336
NM_005927
|
MFAP3
|
microfibrillar-associated protein 3
|
|
chr12_+_51318743
|
0.562
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr1_-_151345156
|
0.562
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr11_+_114310107
|
0.561
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr11_-_104034685
|
0.559
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr11_+_66384052
|
0.559
|
NM_001198845
NM_001198846
NM_001198836
NM_001198837
NM_006328
|
RBM14-RBM4
RBM14
|
RBM14-RBM4 readthrough
RNA binding motif protein 14
|
|
chr7_-_20826503
|
0.557
|
NM_182700
NM_198956
|
SP8
|
Sp8 transcription factor
|
|
chr13_+_49550911
|
0.557
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr9_-_74979625
|
0.555
|
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr1_-_205290756
|
0.553
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr1_+_90024271
|
0.553
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr7_-_94285477
|
0.550
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr13_+_49550627
|
0.545
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr8_-_127570710
|
0.545
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr7_+_114562140
|
0.545
|
NM_001166345
NM_001166346
NM_199072
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr7_+_94285736
|
0.545
|
|
PEG10
|
paternally expressed 10
|
|
chr12_-_110011286
|
0.543
|
NM_052845
|
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type
|
|
chr2_-_145090028
|
0.540
|
|
GTDC1
|
glycosyltransferase-like domain containing 1
|