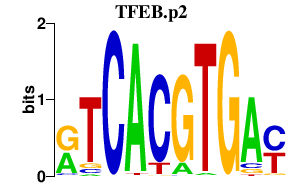
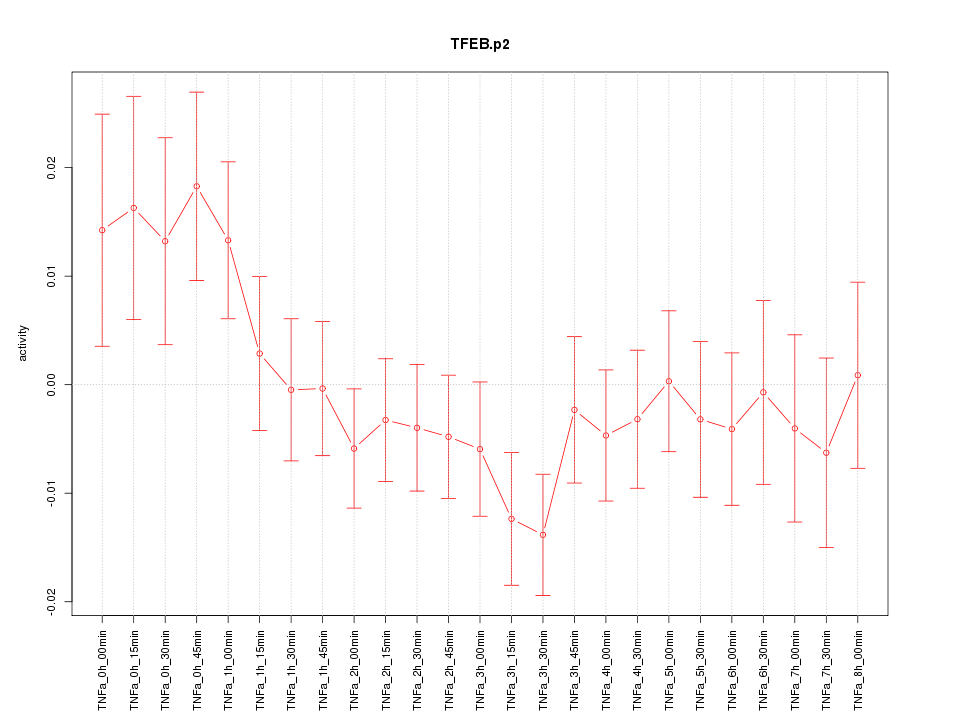
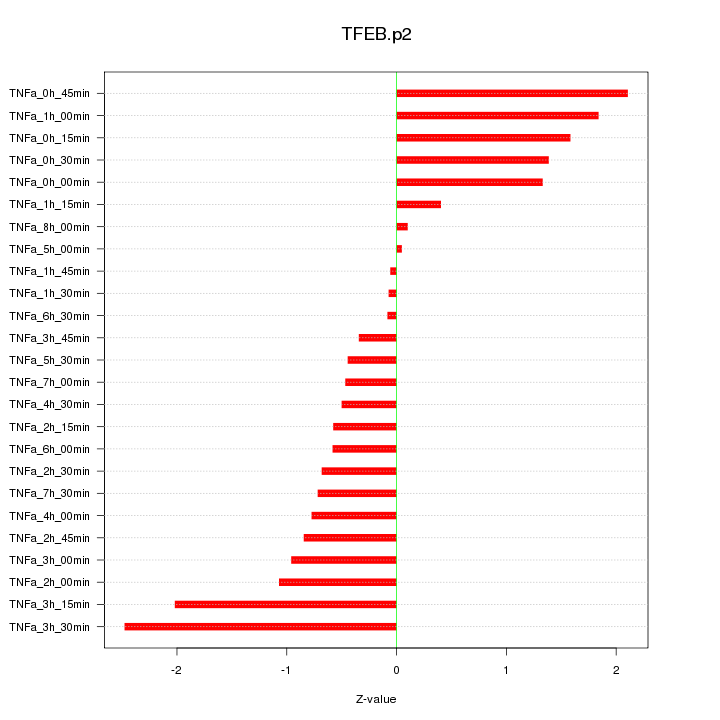
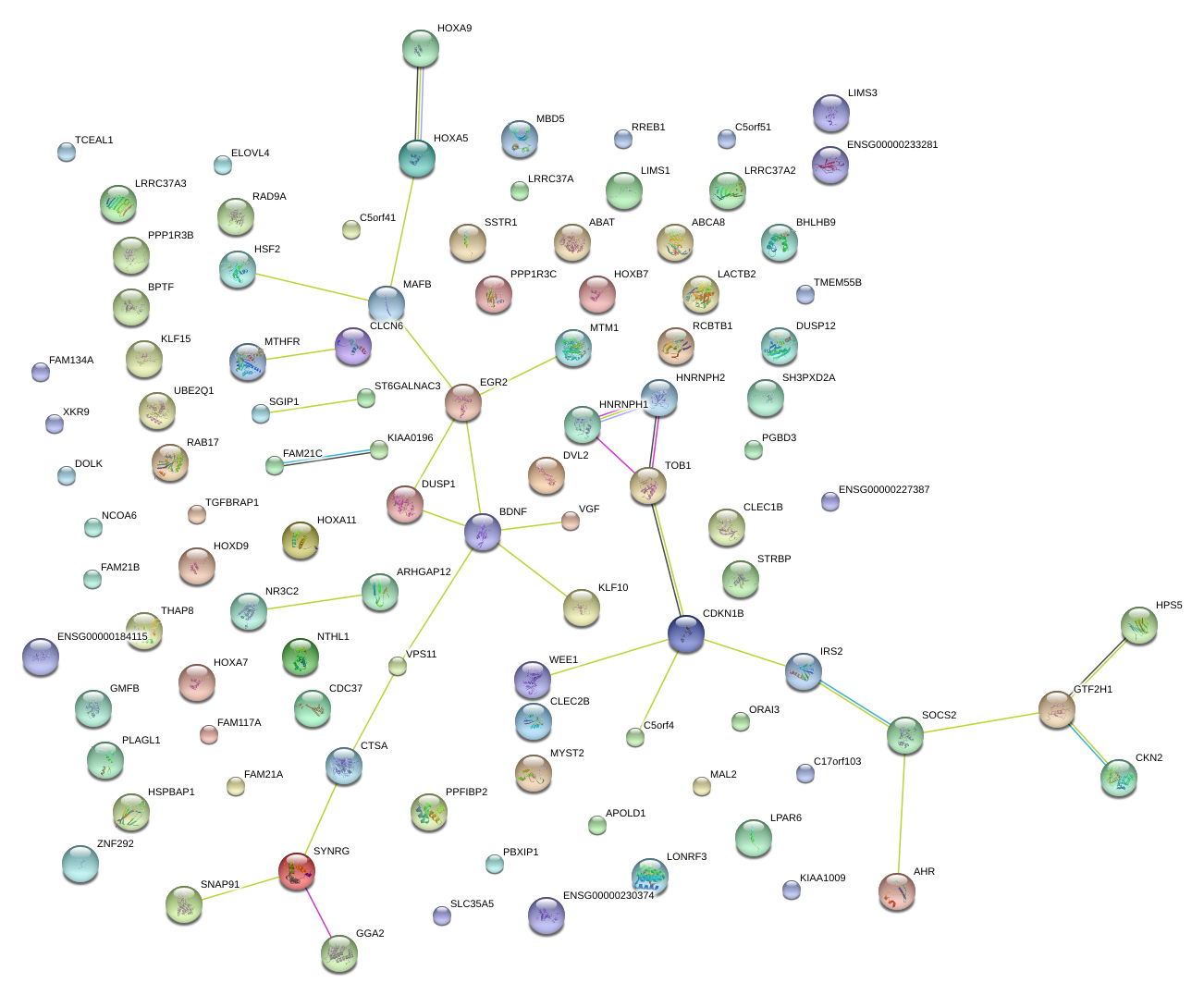
|
chr1_-_11866079
|
7.361
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr7_-_27183225
|
4.889
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chrX_+_101975641
|
4.289
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr17_-_46688362
|
3.852
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr6_+_87865261
|
3.835
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr19_-_36545119
|
3.650
|
|
THAP8
|
THAP domain containing 8
|
|
chr20_-_39317875
|
3.617
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr11_+_67159421
|
3.521
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr10_-_105452878
|
3.467
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr11_+_7535213
|
3.426
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr17_-_47841517
|
3.337
|
NM_030802
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr3_-_122512526
|
3.254
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr5_-_172198160
|
3.232
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr17_-_46688279
|
3.195
|
|
HOXB7
|
homeobox B7
|
|
chr1_-_11865963
|
3.120
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr16_+_30960377
|
3.081
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr17_+_47866061
|
2.997
|
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr7_-_27196163
|
2.985
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr16_+_30960413
|
2.981
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr17_+_47865980
|
2.980
|
NM_001199155
NM_001199156
NM_001199157
NM_001199158
NM_007067
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr5_+_172483285
|
2.952
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr16_+_30960434
|
2.888
|
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr8_-_9009050
|
2.831
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr7_-_27205123
|
2.811
|
|
HOXA9
|
homeobox A9
|
|
chr14_+_38677186
|
2.791
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr10_-_32217687
|
2.768
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr17_-_47841468
|
2.742
|
|
FAM117A
|
family with sequence similarity 117, member A
|
|
chr6_-_84937313
|
2.725
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr6_-_84418705
|
2.715
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr10_-_93392857
|
2.680
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr8_-_71581354
|
2.671
|
|
LACTB2
|
lactamase, beta 2
|
|
chr7_-_27205148
|
2.646
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr10_-_64576123
|
2.643
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr19_-_36545298
|
2.632
|
|
THAP8
|
THAP domain containing 8
|
|
chr16_+_8806825
|
2.609
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr3_+_112280871
|
2.608
|
NM_017945
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr17_-_48945338
|
2.575
|
NM_001243877
|
TOB1
|
transducer of ERBB2, 1
|
|
chr17_+_47866119
|
2.574
|
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr7_-_27224762
|
2.556
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr1_+_11866240
|
2.527
|
|
CLCN6
|
chloride channel 6
|
|
chr10_-_32217762
|
2.513
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr4_-_149363498
|
2.469
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr5_-_154230157
|
2.461
|
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chrX_+_118108575
|
2.448
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr8_+_120220554
|
2.359
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr5_+_41904459
|
2.353
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr2_+_148778573
|
2.344
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr17_+_65821625
|
2.314
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr5_-_154230212
|
2.277
|
NM_032385
|
C5orf4
|
chromosome 5 open reading frame 4
|
|
chr10_-_64576074
|
2.253
|
|
EGR2
|
early growth response 2
|
|
chr13_-_50159628
|
2.237
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr8_-_71581419
|
2.237
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr17_-_35969401
|
2.216
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr1_-_154531083
|
2.203
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr14_-_54955664
|
2.178
|
|
GMFB
|
glia maturation factor, beta
|
|
chr13_-_49001042
|
2.124
|
NM_001162497
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr12_+_12878718
|
2.120
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr14_-_54955714
|
2.113
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chrX_+_118108595
|
2.100
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr6_-_80656870
|
2.078
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr17_-_7137675
|
2.074
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr11_+_118938528
|
2.048
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr17_-_46687925
|
2.044
|
|
HOXB7
|
homeobox B7
|
|
chr7_+_17338182
|
2.031
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr8_+_71581599
|
2.015
|
NM_001011720
|
XKR9
|
XK, Kell blood group complex subunit-related family, member 9
|
|
chr11_+_118938530
|
1.987
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr17_-_21156577
|
1.978
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr11_+_118938515
|
1.972
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr11_-_27723061
|
1.967
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_-_10021932
|
1.955
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr11_-_18343670
|
1.881
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr2_+_220042986
|
1.869
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr8_-_103667882
|
1.843
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr14_-_20929635
|
1.819
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr10_+_46222647
|
1.803
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
|
family with sequence similarity 21, member C
|
|
chr3_+_112280911
|
1.790
|
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr10_-_50747080
|
1.788
|
NM_000124
|
ERCC6
PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6
piggyBac transposable element derived 3
|
|
chr17_-_7137722
|
1.724
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr10_-_50747038
|
1.710
|
|
ERCC6
|
excision repair cross-complementing rodent repair deficiency, complementation group 6
|
|
chrX_+_149737019
|
1.709
|
NM_000252
|
MTM1
|
myotubularin 1
|
|
chr2_+_220042948
|
1.676
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr16_-_2097818
|
1.655
|
NM_002528
|
NTHL1
|
nth endonuclease III-like 1 (E. coli)
|
|
chr13_-_110438896
|
1.652
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr17_-_62915580
|
1.651
|
|
LRRC37A3
|
leucine rich repeat containing 37, member A3
|
|
chr7_-_100808832
|
1.649
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr1_+_76540374
|
1.631
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr8_-_126103926
|
1.631
|
|
KIAA0196
|
KIAA0196
|
|
chr1_+_66999805
|
1.622
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chrX_+_149737083
|
1.616
|
|
MTM1
|
myotubularin 1
|
|
chr6_-_144329342
|
1.607
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr9_-_126030854
|
1.597
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr8_-_126104004
|
1.597
|
|
KIAA0196
|
KIAA0196
|
|
chr20_+_44520021
|
1.592
|
|
CTSA
|
cathepsin A
|
|
chr8_-_126104021
|
1.590
|
|
KIAA0196
|
KIAA0196
|
|
chr9_-_131709783
|
1.579
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr2_-_105946104
|
1.538
|
NM_001142621
NM_004257
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1
|
|
chr9_-_126030792
|
1.532
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr8_-_126103873
|
1.517
|
|
KIAA0196
|
KIAA0196
|
|
chrX_+_102883891
|
1.514
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr3_-_126076057
|
1.513
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr17_-_66951381
|
1.502
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr2_-_238499317
|
1.499
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr6_+_7107986
|
1.487
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr2_+_220042901
|
1.478
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr11_+_9595309
|
1.476
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr1_-_154928595
|
1.464
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr2_+_109204745
|
1.456
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr12_+_93964801
|
1.446
|
|
SOCS2
|
suppressor of cytokine signaling 2
|
|
chr11_+_18344113
|
1.428
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr6_+_122720685
|
1.420
|
NM_001135564
NM_001243094
NM_004506
|
HSF2
|
heat shock transcription factor 2
|
|
chr12_+_12869818
|
1.419
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr16_-_23521755
|
1.405
|
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr1_+_11866206
|
1.392
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr6_+_122720750
|
1.390
|
|
HSF2
|
heat shock transcription factor 2
|
|
chr14_-_20929484
|
1.386
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chrX_+_100663205
|
1.369
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr13_+_42846255
|
1.364
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chrX_-_100548044
|
1.354
|
NM_024885
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa
|
|
chr8_-_95274520
|
1.353
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr8_-_126104045
|
1.349
|
NM_014846
|
KIAA0196
|
KIAA0196
|
|
chr10_+_51827654
|
1.348
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr4_-_99850280
|
1.346
|
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr11_+_9595183
|
1.333
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr11_+_118938473
|
1.332
|
NM_021729
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr5_+_122110762
|
1.328
|
|
SNX2
|
sorting nexin 2
|
|
chrX_+_77166178
|
1.319
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr10_+_51827710
|
1.313
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr12_+_122064448
|
1.286
|
NM_032790
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1
|
|
chr6_-_158589289
|
1.282
|
NM_032861
|
SERAC1
|
serine active site containing 1
|
|
chrX_-_106960268
|
1.271
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr2_+_233562031
|
1.260
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr19_-_41256371
|
1.244
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr5_-_55529185
|
1.241
|
NM_024669
|
ANKRD55
|
ankyrin repeat domain 55
|
|
chr16_-_67184822
|
1.232
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr12_-_122750954
|
1.226
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr19_+_35760002
|
1.214
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr8_-_95274540
|
1.211
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr8_+_74903563
|
1.210
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr5_+_122110778
|
1.208
|
|
SNX2
|
sorting nexin 2
|
|
chr5_+_131892556
|
1.203
|
NM_005732
|
RAD50
|
RAD50 homolog (S. cerevisiae)
|
|
chr12_+_2986361
|
1.194
|
NM_001252499
NM_001252500
|
C12orf32
|
chromosome 12 open reading frame 32
|
|
chr1_+_10092990
|
1.192
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B
|
|
chr7_+_155089614
|
1.188
|
|
INSIG1
|
insulin induced gene 1
|
|
chr5_+_122110737
|
1.188
|
NM_003100
|
SNX2
|
sorting nexin 2
|
|
chrX_+_77166223
|
1.186
|
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr11_+_9595451
|
1.186
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr4_-_76439479
|
1.181
|
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr11_+_9595429
|
1.177
|
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr16_+_5083671
|
1.174
|
|
LOC100507589
|
uncharacterized LOC100507589
|
|
chr4_+_1340957
|
1.169
|
NM_020894
|
KIAA1530
|
KIAA1530
|
|
chr18_-_46987016
|
1.166
|
NM_017653
|
DYM
|
dymeclin
|
|
chr1_-_154928564
|
1.165
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr2_+_177053306
|
1.156
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr6_-_151773168
|
1.150
|
NM_017909
|
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae)
|
|
chr7_+_27225009
|
1.147
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr14_-_39901619
|
1.143
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chrX_-_34675366
|
1.127
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr8_-_54755821
|
1.125
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr5_-_141392537
|
1.115
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr8_-_54755546
|
1.111
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr6_-_31865455
|
1.107
|
NM_006709
NM_025256
|
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2
|
|
chr3_-_4508955
|
1.103
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr3_+_158519888
|
1.099
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr8_-_126104007
|
1.081
|
|
KIAA0196
|
KIAA0196
|
|
chr3_-_134204809
|
1.078
|
NM_001242375
|
ANAPC13
|
anaphase promoting complex subunit 13
|
|
chr17_-_1733083
|
1.076
|
NM_052928
|
SMYD4
|
SET and MYND domain containing 4
|
|
chr2_-_183903225
|
1.076
|
NM_013436
NM_205842
|
NCKAP1
|
NCK-associated protein 1
|
|
chr6_+_64345726
|
1.070
|
|
PHF3
|
PHD finger protein 3
|
|
chr11_+_7535000
|
1.063
|
NM_003621
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr8_-_54755836
|
1.061
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr2_+_109204777
|
1.060
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr17_-_38256905
|
1.058
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr19_-_17375522
|
1.058
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr2_+_176987412
|
1.056
|
NM_014213
|
HOXD9
|
homeobox D9
|
|
chr11_-_57283145
|
1.051
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr3_-_134204840
|
1.047
|
NM_015391
NM_001242374
|
ANAPC13
|
anaphase promoting complex subunit 13
|
|
chr7_-_30029284
|
1.045
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr12_+_72233486
|
1.045
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr3_+_158519911
|
1.043
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr16_-_70719854
|
1.036
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr6_+_42018250
|
1.031
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr6_+_123110345
|
1.026
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr1_-_28969516
|
1.021
|
NM_001135218
NM_005644
|
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa
|
|
chr13_-_52027133
|
1.015
|
|
INTS6
|
integrator complex subunit 6
|
|
chr10_-_46090191
|
1.015
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr17_-_57184114
|
1.004
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr11_+_9685650
|
0.999
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr14_+_74551655
|
0.999
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr3_-_4508939
|
0.996
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr2_+_233561997
|
0.995
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr4_-_47465719
|
0.988
|
|
COMMD8
|
COMM domain containing 8
|
|
chr2_-_62733475
|
0.977
|
NM_198276
|
TMEM17
|
transmembrane protein 17
|
|
chr7_-_14031049
|
0.970
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr17_+_30677187
|
0.960
|
|
ZNF207
|
zinc finger protein 207
|
|
chr4_-_76439582
|
0.953
|
NM_001008925
NM_001009922
NM_015436
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chr12_-_110434187
|
0.947
|
NM_001135213
NM_001135214
NM_014776
NM_057169
NM_057170
NM_139201
|
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2
|
|
chr8_-_82754427
|
0.942
|
NM_022133
NM_152836
NM_152837
|
SNX16
|
sorting nexin 16
|
|
chr11_+_9685624
|
0.940
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr16_-_71318761
|
0.935
|
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr17_+_42977141
|
0.934
|
NM_213607
|
CCDC103
|
coiled-coil domain containing 103
|