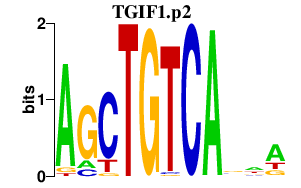
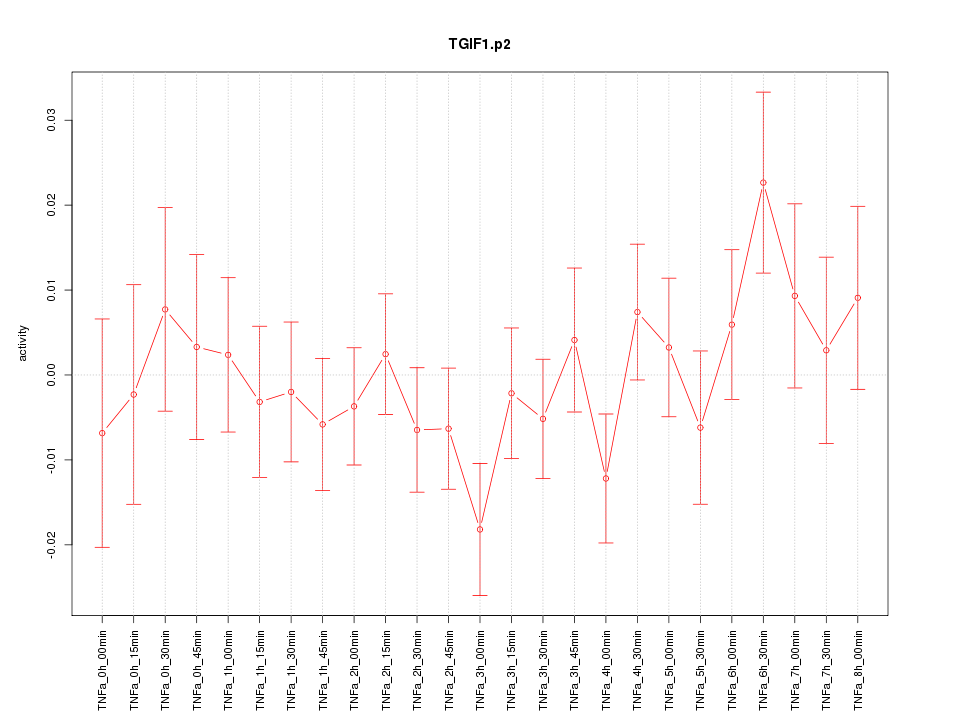
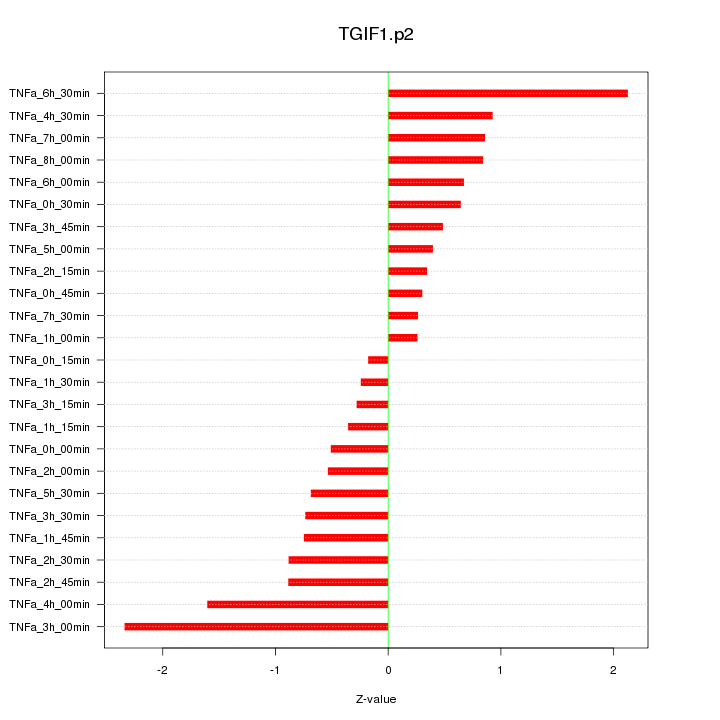
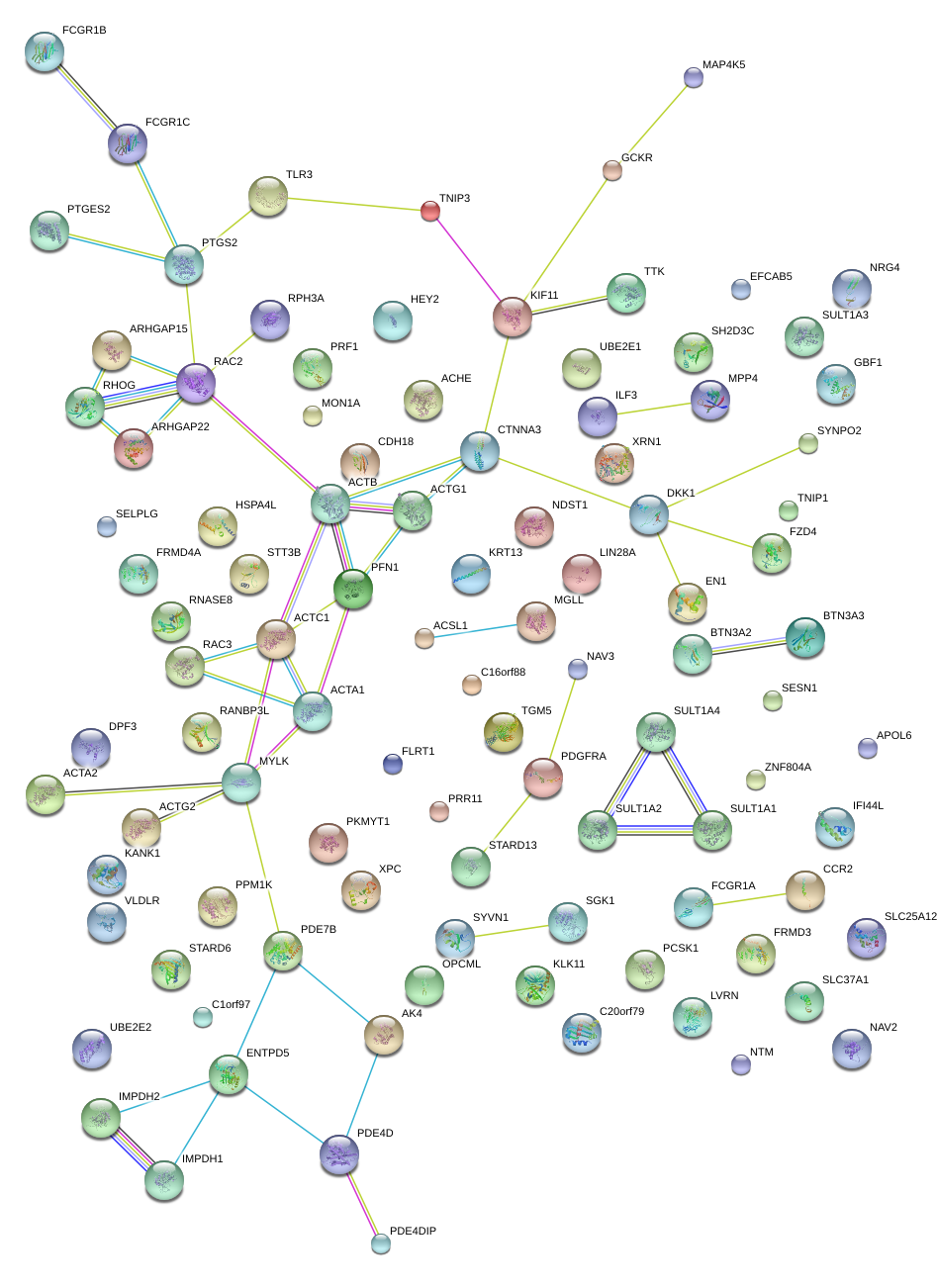
|
chr1_+_79086087
|
3.454
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr3_-_123339178
|
2.156
|
|
MYLK
|
myosin light chain kinase
|
|
chr12_+_78225068
|
1.464
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr22_+_36044412
|
1.375
|
NM_030641
|
APOL6
|
apolipoprotein L, 6
|
|
chr5_-_150466696
|
1.235
|
NM_001252391
NM_001252392
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr3_-_127541975
|
1.215
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr13_-_34250899
|
1.208
|
NM_001243476
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_-_123339423
|
1.191
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr3_-_127541160
|
1.175
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr5_-_58571916
|
1.169
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_-_134639195
|
1.168
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr4_+_186990308
|
1.134
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr1_-_186649421
|
1.072
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr9_-_85882093
|
0.988
|
NM_001244962
|
FRMD3
|
FERM domain containing 3
|
|
chr5_-_19988293
|
0.985
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr6_+_80714389
|
0.943
|
|
TTK
|
TTK protein kinase
|
|
chr9_+_127420690
|
0.917
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr5_-_58882274
|
0.909
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr13_-_33859835
|
0.883
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_-_86666211
|
0.879
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr5_+_115298136
|
0.857
|
NM_173800
|
AQPEP
|
laeverin
|
|
chr6_+_80714321
|
0.836
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr3_+_31574431
|
0.830
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr10_-_90750955
|
0.780
|
NM_001141945
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr3_+_31574119
|
0.776
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr11_+_19734821
|
0.772
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr14_+_21525980
|
0.728
|
NM_138331
|
RNASE8
|
ribonuclease, RNase A family, 8
|
|
chr11_+_63871240
|
0.703
|
NM_013280
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr5_-_36301999
|
0.690
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr2_+_143886898
|
0.685
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr10_-_69455926
|
0.682
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr11_+_131240370
|
0.680
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr10_-_14372807
|
0.678
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_+_65613771
|
0.674
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr2_+_143886969
|
0.672
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr20_+_18794369
|
0.657
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr15_-_43559054
|
0.645
|
NM_004245
NM_201631
|
TGM5
|
transglutaminase 5
|
|
chr4_-_122137781
|
0.642
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr5_-_150467220
|
0.629
|
NM_001252390
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr4_-_185747051
|
0.627
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr4_+_55095263
|
0.622
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr2_+_27719702
|
0.617
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr2_-_202563353
|
0.613
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr3_+_23244703
|
0.611
|
NM_152653
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2
|
|
chr17_-_39661803
|
0.610
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr4_-_122085475
|
0.605
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr3_+_23244486
|
0.595
|
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2
|
|
chr18_-_51880942
|
0.595
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr1_-_120935893
|
0.584
|
|
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr4_-_89205667
|
0.567
|
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K
|
|
chr10_+_104005307
|
0.565
|
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr12_+_113229548
|
0.564
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr21_+_43933983
|
0.564
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr1_+_65613211
|
0.558
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr1_-_145039631
|
0.554
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr4_+_128703402
|
0.543
|
NM_014278
|
HSPA4L
|
heat shock 70kDa protein 4-like
|
|
chr16_+_30210548
|
0.534
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr7_-_100493481
|
0.533
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr3_+_46395574
|
0.529
|
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr10_-_49812996
|
0.525
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr4_+_55095433
|
0.515
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr22_-_37640287
|
0.510
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr13_-_60325073
|
0.505
|
|
|
|
|
chr17_-_4851797
|
0.492
|
NM_005022
|
PFN1
|
profilin 1
|
|
chr3_-_49967194
|
0.486
|
NM_001142501
NM_032355
|
MON1A
|
MON1 homolog A (yeast)
|
|
chr3_-_49066858
|
0.486
|
NM_000884
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr9_-_130540911
|
0.484
|
NM_170600
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr17_-_4851827
|
0.483
|
|
PFN1
|
profilin 1
|
|
chr16_+_29471206
|
0.483
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr17_+_57232859
|
0.482
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr17_-_4851688
|
0.479
|
|
PFN1
|
profilin 1
|
|
chr6_+_126070895
|
0.478
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr4_+_128703480
|
0.478
|
|
HSPA4L
|
heat shock 70kDa protein 4-like
|
|
chr16_-_3030473
|
0.472
|
NM_004203
NM_182687
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr5_-_58652671
|
0.469
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_-_109415514
|
0.468
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr9_+_2622099
|
0.463
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr14_-_73360792
|
0.460
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr9_+_2622134
|
0.456
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr7_+_21468293
|
0.456
|
|
|
|
|
chr12_-_109027654
|
0.456
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr1_+_26737232
|
0.455
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr3_-_14220071
|
0.451
|
NM_001145769
NM_004628
|
XPC
|
xeroderma pigmentosum, complementation group C
|
|
chr10_+_94352960
|
0.450
|
|
KIF11
|
kinesin family member 11
|
|
chr4_+_119948554
|
0.445
|
|
SYNPO2
|
synaptopodin 2
|
|
chr9_+_2622052
|
0.440
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr5_-_95748688
|
0.439
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr9_+_706744
|
0.437
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr19_-_51530781
|
0.436
|
NM_144947
|
KLK11
|
kallikrein-related peptidase 11
|
|
chr1_+_65613971
|
0.434
|
|
AK4
|
adenylate kinase 4
|
|
chr3_-_142166782
|
0.433
|
NM_001042604
NM_019001
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr2_-_172750724
|
0.431
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr10_+_54074037
|
0.428
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr12_-_109027565
|
0.428
|
|
SELPLG
|
selectin P ligand
|
|
chr2_-_119605758
|
0.422
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr2_+_185463235
|
0.421
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr11_-_64901995
|
0.420
|
NM_032431
NM_172230
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin
|
|
chr6_+_26368405
|
0.418
|
NM_001197246
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr11_-_64901943
|
0.417
|
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin
|
|
chr15_-_76352068
|
0.408
|
|
NRG4
|
neuregulin 4
|
|
chr17_+_28256873
|
0.407
|
NM_001145053
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr3_-_49066813
|
0.407
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr5_+_149865310
|
0.402
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr6_+_136172802
|
0.398
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_+_185463092
|
0.397
|
NM_194250
|
ZNF804A
|
zinc finger protein 804A
|
|
chr10_+_104005254
|
0.395
|
NM_001199378
NM_001199379
NM_004193
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1
|
|
chr13_+_27844463
|
0.394
|
NM_206827
|
RASL11A
|
RAS-like, family 11, member A
|
|
chr2_+_167744996
|
0.388
|
NM_001079810
NM_001199143
NM_152381
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr13_+_108922560
|
0.387
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chrX_-_63450486
|
0.385
|
NM_130388
|
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chr3_-_49066760
|
0.380
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr10_+_98592658
|
0.380
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr18_+_61554938
|
0.377
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr22_+_46692637
|
0.377
|
NM_016426
|
GTSE1
|
G-2 and S-phase expressed 1
|
|
chr19_-_14228362
|
0.371
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr15_-_72522062
|
0.368
|
NM_001206799
|
PKM2
|
pyruvate kinase, muscle
|
|
chr1_+_203097427
|
0.366
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr3_+_67048726
|
0.360
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr6_-_41130921
|
0.358
|
NM_018965
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr11_-_57195052
|
0.354
|
NM_017611
|
SLC43A3
|
solute carrier family 43, member 3
|
|
chr2_+_97202463
|
0.350
|
NM_212481
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like)
|
|
chr8_-_145060594
|
0.349
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr6_+_31913852
|
0.347
|
|
CFB
|
complement factor B
|
|
chr3_-_10332408
|
0.346
|
NM_001134941
NM_016362
|
GHRL
|
ghrelin/obestatin prepropeptide
|
|
chr5_+_133984234
|
0.345
|
NM_001252231
NM_021982
|
SEC24A
|
SEC24 family, member A (S. cerevisiae)
|
|
chr11_+_73661363
|
0.344
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr16_+_57673206
|
0.343
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr7_+_101928390
|
0.341
|
NM_020979
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr2_+_242127923
|
0.341
|
NM_001001666
NM_001001891
|
ANO7
|
anoctamin 7
|
|
chr8_+_80523048
|
0.340
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr2_-_176866925
|
0.339
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chrX_-_128788913
|
0.339
|
NM_017413
|
APLN
|
apelin
|
|
chr5_-_146833150
|
0.339
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr7_+_101928417
|
0.338
|
|
SH2B2
|
SH2B adaptor protein 2
|
|
chrX_-_153775774
|
0.338
|
NM_001042351
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chrX_+_153775561
|
0.335
|
NM_001099857
NM_001145255
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr11_-_8954534
|
0.335
|
NM_020643
|
C11orf16
|
chromosome 11 open reading frame 16
|
|
chr17_+_13972841
|
0.335
|
|
COX10
|
COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast)
|
|
chr7_-_97501680
|
0.334
|
|
ASNS
|
asparagine synthetase (glutamine-hydrolyzing)
|
|
chr17_+_57233100
|
0.334
|
|
PRR11
|
proline rich 11
|
|
chr20_+_8112762
|
0.334
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr10_+_69869249
|
0.334
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr9_-_35115844
|
0.333
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr16_+_31366519
|
0.332
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr18_+_21693316
|
0.332
|
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr19_-_39235113
|
0.330
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr5_-_169724782
|
0.330
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr11_-_57004674
|
0.328
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chr8_-_134115309
|
0.328
|
NM_001045556
NM_001045557
|
SLA
|
Src-like-adaptor
|
|
chr10_+_60028885
|
0.328
|
NM_018464
|
CISD1
|
CDGSH iron sulfur domain 1
|
|
chr18_+_32073343
|
0.326
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr8_-_15095791
|
0.326
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chr4_-_123542165
|
0.324
|
NM_001207006
NM_021803
|
IL21
|
interleukin 21
|
|
chr4_+_71494460
|
0.323
|
NM_031889
|
ENAM
|
enamelin
|
|
chr1_+_39875175
|
0.322
|
NM_015038
|
KIAA0754
|
KIAA0754
|
|
chr5_+_173472737
|
0.321
|
|
HMP19
|
HMP19 protein
|
|
chr11_-_568419
|
0.320
|
|
MIR210HG
|
MIR210 host gene (non-protein coding)
|
|
chrX_-_154563735
|
0.318
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr13_+_108922205
|
0.316
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr12_-_109027615
|
0.312
|
|
SELPLG
|
selectin P ligand
|
|
chr4_-_89205887
|
0.308
|
NM_152542
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K
|
|
chr6_-_119031230
|
0.307
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr2_+_220436953
|
0.306
|
NM_002191
|
INHA
|
inhibin, alpha
|
|
chr16_-_28621279
|
0.305
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr17_+_16945766
|
0.305
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr7_-_97501737
|
0.304
|
NM_001178075
NM_001178076
NM_001673
NM_183356
|
ASNS
|
asparagine synthetase (glutamine-hydrolyzing)
|
|
chr17_+_41857802
|
0.301
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chrX_+_153776061
|
0.299
|
NM_003639
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr5_-_41071404
|
0.298
|
NM_173489
|
HEATR7B2
|
HEAT repeat family member 7B2
|
|
chr4_-_185747183
|
0.296
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr3_-_122283423
|
0.292
|
NM_001146103
NM_031458
|
PARP9
|
poly (ADP-ribose) polymerase family, member 9
|
|
chr3_+_178254223
|
0.291
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr17_-_39525873
|
0.290
|
NM_002279
|
KRT33B
|
keratin 33B
|
|
chr12_-_25348031
|
0.290
|
NM_001082972
NM_001082973
NM_001204101
NM_001204102
NM_018272
|
CASC1
|
cancer susceptibility candidate 1
|
|
chr12_+_124773709
|
0.289
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr10_+_91498724
|
0.289
|
|
KIF20B
|
kinesin family member 20B
|
|
chr1_-_44482996
|
0.286
|
NM_006934
NM_201649
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr3_-_183273407
|
0.286
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr9_-_117554809
|
0.285
|
NM_001204344
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr6_+_37225506
|
0.285
|
NM_017772
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr6_+_29274402
|
0.284
|
NM_030946
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1
|
|
chr16_-_55989942
|
0.282
|
NM_001190158
|
CES5A
|
carboxylesterase 5A
|
|
chr5_-_64777703
|
0.282
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr17_-_45933138
|
0.282
|
NM_199262
|
SP6
|
Sp6 transcription factor
|
|
chr8_-_145060563
|
0.280
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr8_-_103251002
|
0.279
|
NM_001172477
NM_001172478
NM_015713
|
RRM2B
|
ribonucleotide reductase M2 B (TP53 inducible)
|
|
chr2_-_100721792
|
0.279
|
NM_001025108
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr6_-_41130820
|
0.276
|
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr20_+_54987167
|
0.275
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr17_-_39341126
|
0.274
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr11_+_35198199
|
0.274
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr16_+_88636841
|
0.273
|
|
ZC3H18
|
zinc finger CCCH-type containing 18
|
|
chr12_-_125473618
|
0.273
|
NM_032656
|
DHX37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37
|
|
chr12_-_51740422
|
0.272
|
NM_001971
|
CELA1
|
chymotrypsin-like elastase family, member 1
|
|
chr22_-_37640187
|
0.271
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr5_-_75008245
|
0.271
|
NM_152408
|
POC5
|
POC5 centriolar protein homolog (Chlamydomonas)
|
|
chr7_-_130371305
|
0.270
|
NM_052933
|
TSGA13
|
testis specific, 13
|
|
chr22_-_36013303
|
0.270
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chrX_+_153775828
|
0.268
|
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr22_-_39052579
|
0.268
|
NM_001013647
|
LOC646851
|
putative uncharacterized protein LOC388900
|