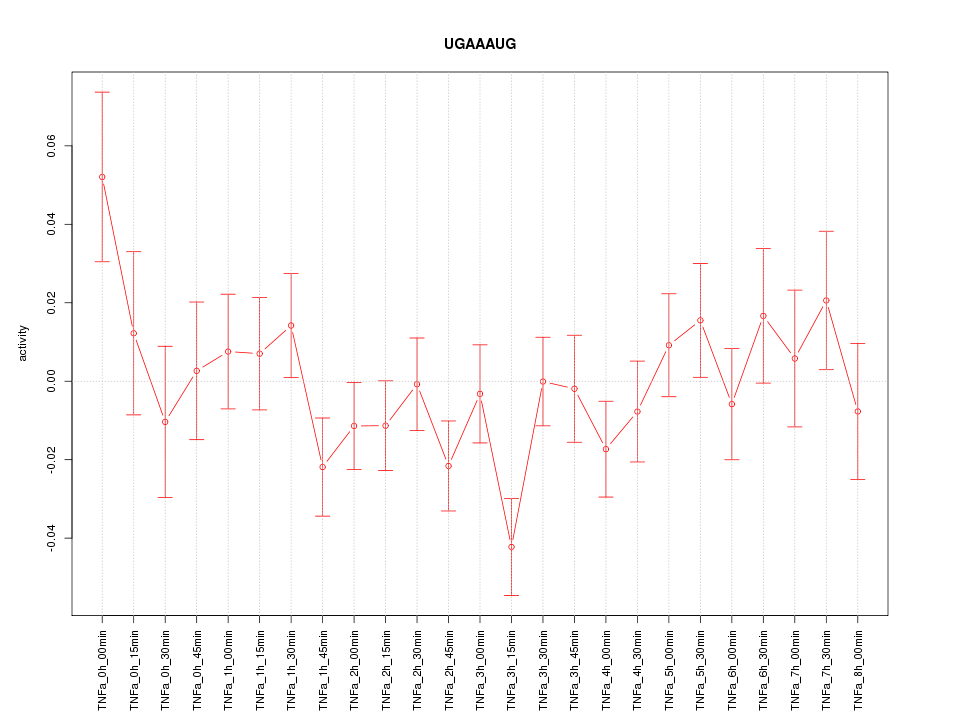
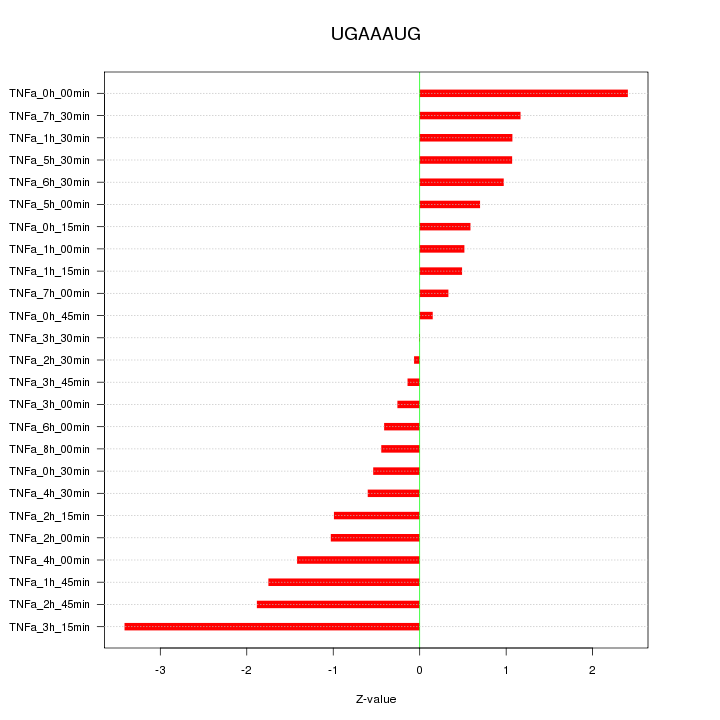
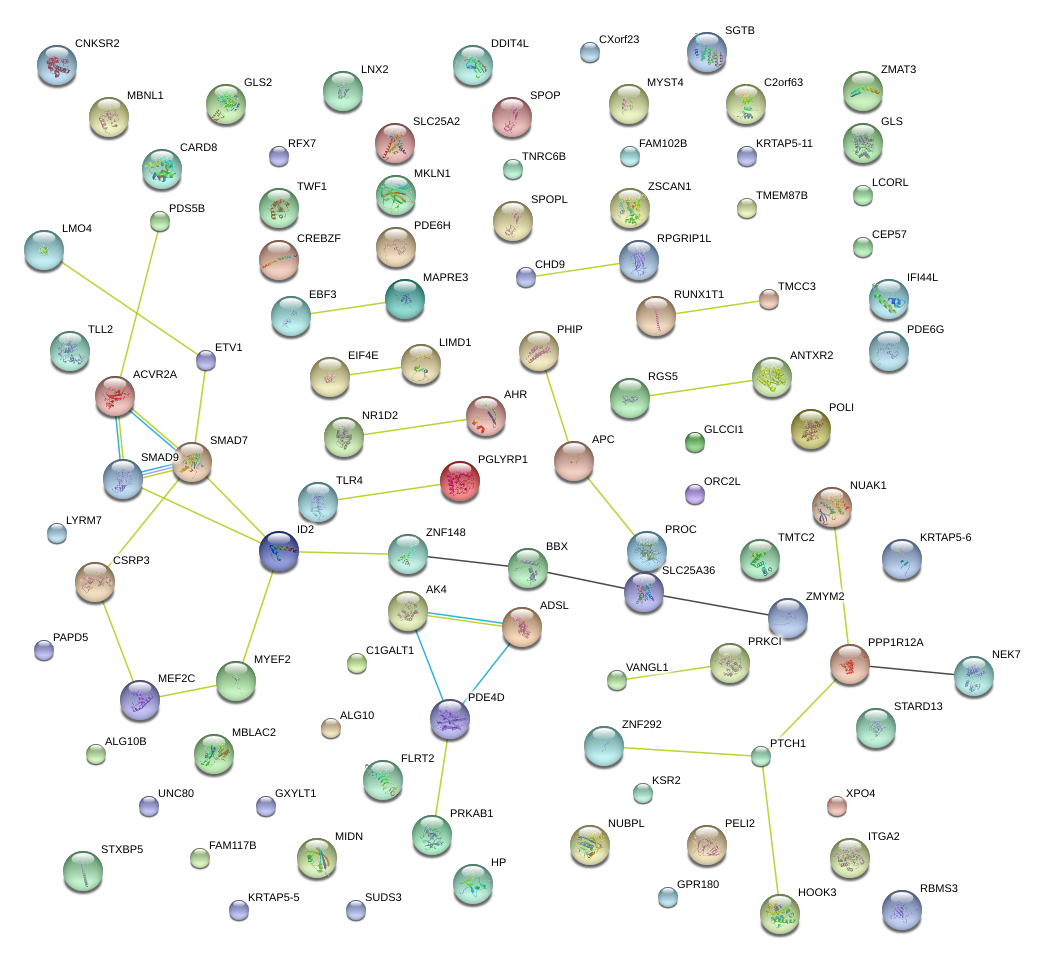
|
chr12_-_95044205
|
0.933
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr5_+_52284903
|
0.876
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr7_+_17338182
|
0.871
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr3_+_107241782
|
0.857
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr16_+_50186809
|
0.758
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr12_-_106533810
|
0.735
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr13_-_37494370
|
0.715
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr18_-_46477080
|
0.628
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr8_-_93115453
|
0.609
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_185126153
|
0.608
|
NM_001202423
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr13_-_21476895
|
0.604
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr18_-_46475702
|
0.599
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr18_+_51795848
|
0.591
|
NM_007195
|
POLI
|
polymerase (DNA directed) iota
|
|
chr1_-_185125819
|
0.590
|
NM_030934
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr8_-_93029864
|
0.585
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_23987514
|
0.581
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr13_+_20532788
|
0.581
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr8_-_93107881
|
0.580
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107442
|
0.576
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr19_-_48752921
|
0.576
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr18_-_46469118
|
0.573
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr6_+_87865261
|
0.572
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr8_-_93111915
|
0.565
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_80329234
|
0.564
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr6_-_79787939
|
0.563
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr16_-_53737713
|
0.562
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr10_-_131762017
|
0.554
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr12_-_42538432
|
0.553
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr3_-_178789597
|
0.553
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr16_+_53088791
|
0.540
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr2_+_203499819
|
0.535
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr9_+_120466453
|
0.528
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr19_-_48744276
|
0.520
|
NM_001184900
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr14_+_56584854
|
0.513
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr12_+_38710556
|
0.511
|
NM_001013620
|
ALG10B
|
asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog B (yeast)
|
|
chr5_+_112073555
|
0.507
|
NM_000038
NM_001127510
|
APC
|
adenomatous polyposis coli
|
|
chr2_+_112812799
|
0.504
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr13_+_95254021
|
0.501
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr1_+_15250578
|
0.501
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr5_+_112043188
|
0.499
|
NM_001127511
|
APC
|
adenomatous polyposis coli
|
|
chr3_+_23986735
|
0.499
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr12_+_83080901
|
0.492
|
NM_152588
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr8_-_93075190
|
0.491
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_89770432
|
0.490
|
NM_203406
|
MBLAC2
|
metallo-beta-lactamase domain containing 2
|
|
chr1_+_15256295
|
0.490
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr9_-_98269480
|
0.486
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr1_-_163172624
|
0.477
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr12_-_118406027
|
0.471
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr5_-_65017874
|
0.462
|
NM_019072
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr12_-_80307690
|
0.461
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr14_+_85996454
|
0.461
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr12_-_80328926
|
0.460
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr3_+_45636176
|
0.459
|
NM_014240
|
LIMD1
|
LIM domains containing 1
|
|
chr5_-_58334936
|
0.458
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_198125890
|
0.454
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr13_-_28194677
|
0.442
|
NM_153371
|
LNX2
|
ligand of numb-protein X 2
|
|
chr9_-_98270321
|
0.440
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr9_-_98270830
|
0.434
|
NM_000264
|
PTCH1
|
patched 1
|
|
chr4_-_80994309
|
0.432
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr10_+_76586299
|
0.430
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr2_-_55459698
|
0.424
|
NM_001135598
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chr1_-_163172863
|
0.424
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_+_109102970
|
0.421
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr4_-_101111606
|
0.419
|
NM_145244
|
DDIT4L
|
DNA-damage-inducible transcript 4-like
|
|
chr13_+_20532906
|
0.418
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr11_+_95523608
|
0.416
|
NM_001243776
NM_001243777
NM_014679
|
CEP57
|
centrosomal protein 57kDa
|
|
chr2_-_55459406
|
0.414
|
NM_152385
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chr13_-_33780142
|
0.411
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_-_59189620
|
0.410
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_+_7222142
|
0.410
|
NM_020156
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1
|
|
chr13_-_33859835
|
0.408
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr13_-_33760215
|
0.408
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_+_116184573
|
0.408
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr1_+_65613211
|
0.407
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr3_+_29322802
|
0.401
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr12_-_44200047
|
0.400
|
NM_001242397
NM_002822
|
TWF1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr7_+_8008416
|
0.399
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr22_+_40440664
|
0.397
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr19_+_58545399
|
0.396
|
NM_182572
|
ZSCAN1
|
zinc finger and SCAN domain containing 1
|
|
chr5_-_58295758
|
0.392
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_18023358
|
0.391
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr15_-_48470453
|
0.388
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr2_+_148602569
|
0.388
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr3_+_140660639
|
0.386
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr12_+_118814310
|
0.386
|
NM_022491
|
SUDS3
|
suppressor of defective silencing 3 homolog (S. cerevisiae)
|
|
chr1_+_15272414
|
0.385
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chrX_-_19988381
|
0.384
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr5_-_88179278
|
0.382
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_116185249
|
0.381
|
NM_001172412
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr2_+_139259349
|
0.381
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr12_+_120105656
|
0.381
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr4_-_99850242
|
0.381
|
NM_001130678
|
EIF4E
|
eukaryotic translation initiation factor 4E
|
|
chr14_+_32030579
|
0.379
|
NM_025152
|
NUBPL
|
nucleotide binding protein-like
|
|
chr19_+_1248548
|
0.376
|
NM_177401
|
MIDN
|
midnolin
|
|
chr14_+_32047121
|
0.376
|
NM_001201573
|
NUBPL
|
nucleotide binding protein-like
|
|
chr1_+_65613771
|
0.373
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr2_+_210636700
|
0.372
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chr15_-_56535475
|
0.367
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr5_+_130506635
|
0.367
|
NM_181705
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr9_-_98269686
|
0.366
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr3_-_125094003
|
0.366
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr12_+_15125955
|
0.365
|
NM_006205
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma
|
|
chr7_+_130794854
|
0.364
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr3_+_152017193
|
0.360
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr13_+_33160537
|
0.359
|
NM_015032
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr10_-_98273444
|
0.356
|
NM_012465
|
TLL2
|
tolloid-like 2
|
|
chr8_+_42752019
|
0.355
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr6_+_147525493
|
0.354
|
NM_001127715
NM_139244
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr2_+_191745546
|
0.353
|
NM_014905
|
GLS
|
glutaminase
|
|
chr7_+_131012594
|
0.353
|
NM_013255
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr5_-_58652671
|
0.352
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_14031049
|
0.350
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr1_+_87794150
|
0.349
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr11_-_85376159
|
0.345
|
NM_001039618
|
CREBZF
|
CREB/ATF bZIP transcription factor
|
|
chr6_+_96969669
|
0.343
|
NM_015323
|
UFL1
|
UFM1-specific ligase 1
|
|
chr11_+_1718424
|
0.338
|
NM_001012416
|
KRTAP5-6
|
keratin associated protein 5-6
|
|
chr2_+_8822112
|
0.338
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr3_+_169940214
|
0.336
|
NM_002740
|
PRKCI
|
protein kinase C, iota
|
|
chr2_-_201828180
|
0.336
|
NM_006190
|
ORC2
|
origin recognition complex, subunit 2
|
|
chr1_+_79086087
|
0.333
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr15_-_43398168
|
0.333
|
NM_174916
|
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1
|
|
chr1_+_28696056
|
0.333
|
NM_001048183
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr5_+_77656338
|
0.332
|
NM_004866
|
SCAMP1
|
secretory carrier membrane protein 1
|
|
chr12_+_107168336
|
0.332
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr4_-_185747183
|
0.332
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chrX_-_114468604
|
0.332
|
NM_001243963
NM_020871
|
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2
|
|
chr12_-_8088629
|
0.331
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr3_+_151985828
|
0.330
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr6_+_147829784
|
0.327
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr12_+_46123495
|
0.327
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr5_-_79286793
|
0.325
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chr17_+_65821625
|
0.324
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chrX_-_20284708
|
0.322
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr5_-_58571916
|
0.321
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_88199868
|
0.321
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr20_+_31350190
|
0.320
|
NM_001207055
NM_001207056
NM_006892
NM_175848
NM_175849
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr9_-_98279246
|
0.320
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr13_+_39612440
|
0.318
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr9_+_4490193
|
0.316
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr5_-_59783885
|
0.313
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_+_138483024
|
0.311
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr11_-_86666211
|
0.310
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr12_-_26985922
|
0.307
|
NM_002223
|
ITPR2
|
inositol 1,4,5-trisphosphate receptor, type 2
|
|
chr14_+_57857261
|
0.305
|
NM_001011713
|
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit
|
|
chr9_+_4662285
|
0.305
|
NM_203453
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2
|
|
chr14_+_32183774
|
0.303
|
NM_001201574
|
NUBPL
|
nucleotide binding protein-like
|
|
chr11_-_130184459
|
0.302
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr2_-_7005764
|
0.302
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr4_-_79860369
|
0.300
|
NM_001040202
|
PAQR3
|
progestin and adipoQ receptor family member III
|
|
chr14_-_50319498
|
0.300
|
NM_004713
|
NEMF
|
nuclear export mediator factor
|
|
chr20_+_31367637
|
0.297
|
NM_175850
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr19_+_33571785
|
0.297
|
NM_018025
|
GPATCH1
|
G patch domain containing 1
|
|
chr6_+_143771779
|
0.296
|
NM_003630
|
PEX3
|
peroxisomal biogenesis factor 3
|
|
chr7_+_39989955
|
0.294
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr2_-_208890207
|
0.293
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr8_+_95732101
|
0.289
|
NM_181787
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr2_+_112656180
|
0.288
|
NM_006343
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr5_+_148724976
|
0.283
|
NM_152407
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr8_-_133493003
|
0.283
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr5_-_59064437
|
0.279
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_170303774
|
0.276
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (orphan transporter), member 14
|
|
chr10_+_91152321
|
0.276
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr1_+_28764660
|
0.273
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr1_+_112162397
|
0.273
|
NM_001010935
NM_002884
|
RAP1A
|
RAP1A, member of RAS oncogene family
|
|
chr9_-_88969323
|
0.270
|
NM_001185059
NM_001185074
NM_024617
|
ZCCHC6
|
zinc finger, CCHC domain containing 6
|
|
chr19_-_23941571
|
0.269
|
NM_138286
|
ZNF681
|
zinc finger protein 681
|
|
chr10_-_12085022
|
0.268
|
NM_080599
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr10_+_54074037
|
0.267
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr17_-_62658311
|
0.267
|
NM_022739
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr4_-_135122902
|
0.265
|
NM_001114734
|
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like
|
|
chr5_-_64777703
|
0.264
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr7_-_27135524
|
0.263
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr3_+_16216155
|
0.262
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr8_+_67624652
|
0.261
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr19_-_11308184
|
0.261
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr2_-_39664129
|
0.261
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr22_+_39853316
|
0.260
|
NM_002409
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr4_-_166034003
|
0.259
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr4_-_186125072
|
0.259
|
NM_020827
|
KIAA1430
|
KIAA1430
|
|
chr15_+_63481713
|
0.258
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr19_+_38810461
|
0.258
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr2_-_201936389
|
0.258
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr10_+_94449678
|
0.258
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr6_+_143999058
|
0.257
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr18_+_72922725
|
0.256
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr4_-_119757288
|
0.256
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr10_+_92980322
|
0.254
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr8_+_67687415
|
0.254
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr9_+_37800779
|
0.251
|
NM_024345
|
DCAF10
|
DDB1 and CUL4 associated factor 10
|
|
chr2_+_113033146
|
0.250
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr4_+_186317837
|
0.250
|
NM_181726
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr7_+_21467572
|
0.249
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chrX_-_118739812
|
0.249
|
NM_001173487
NM_017544
|
NKRF
|
NFKB repressing factor
|
|
chr19_-_11305186
|
0.249
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr5_-_100238870
|
0.248
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr3_-_148804122
|
0.248
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr8_-_103251002
|
0.248
|
NM_001172477
NM_001172478
NM_015713
|
RRM2B
|
ribonucleotide reductase M2 B (TP53 inducible)
|
|
chr4_+_157997181
|
0.248
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr1_-_205091137
|
0.246
|
NM_001193272
NM_001193273
NM_005057
|
RBBP5
|
retinoblastoma binding protein 5
|
|
chr3_+_143692166
|
0.244
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|