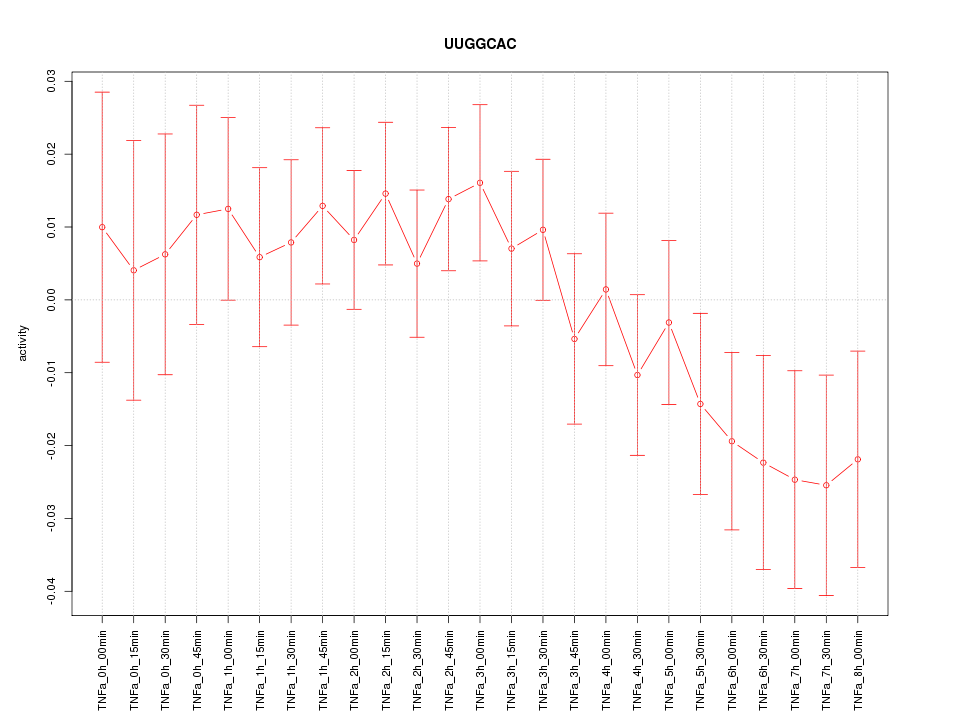
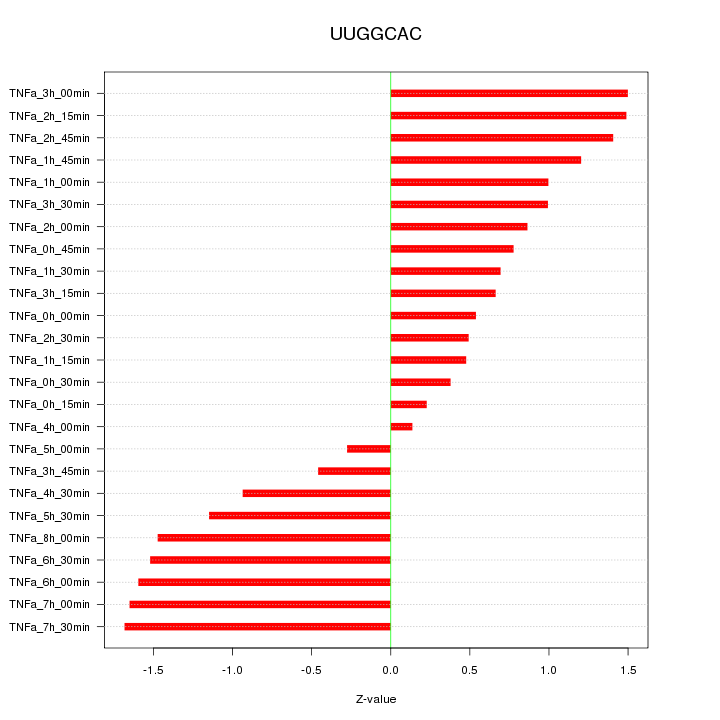
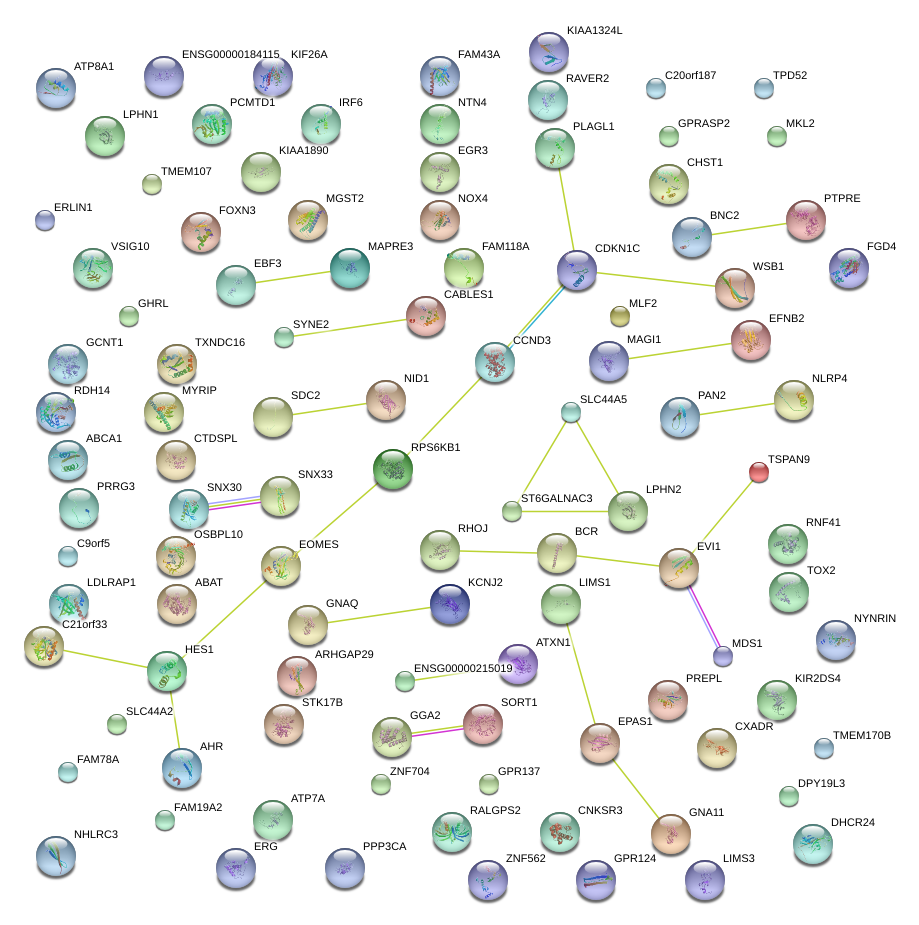
|
chr9_-_107690526
|
4.525
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr9_+_79056581
|
4.378
|
NM_001097634
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr1_-_209979388
|
4.351
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr3_+_39851028
|
4.230
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr9_+_79093256
|
4.117
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79115548
|
4.028
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_+_79074067
|
3.900
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr18_+_20735788
|
3.881
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr18_+_20715726
|
3.841
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr17_+_68165660
|
3.562
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr10_-_131762017
|
3.399
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr8_+_37654395
|
3.077
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr13_-_107187312
|
2.821
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr14_+_64680858
|
2.683
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr8_-_80993009
|
2.667
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr1_+_82266081
|
2.662
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr14_+_64682671
|
2.654
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr8_-_81083660
|
2.611
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr1_-_55352920
|
2.516
|
NM_014762
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr4_-_102268626
|
2.513
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr12_-_96184510
|
2.407
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr21_+_18885104
|
2.374
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr3_+_193853930
|
2.357
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr20_+_42544781
|
2.317
|
NM_001098796
NM_032883
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr9_+_115513050
|
2.291
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr7_-_86595203
|
2.269
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr9_-_134151905
|
2.269
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr8_-_22549901
|
2.239
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr8_-_22550708
|
2.230
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr20_+_42574535
|
2.197
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr3_+_37903643
|
2.156
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr6_-_16761600
|
2.141
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr8_-_22550057
|
2.122
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr9_-_80646150
|
2.065
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr20_+_42543417
|
2.044
|
NM_001098797
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr3_-_66024508
|
2.003
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chrX_+_101967103
|
1.994
|
NM_001004051
NM_001184874
NM_001184875
NM_001184876
NM_138437
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr1_+_65210724
|
1.876
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr7_-_86688944
|
1.862
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr8_-_81787015
|
1.838
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr12_+_32654976
|
1.830
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr12_-_56615679
|
1.822
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr11_-_2906960
|
1.798
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr12_-_56615484
|
1.793
|
NM_001242826
|
RNF41
|
ring finger protein 41
|
|
chrX_+_77166178
|
1.740
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr2_+_46524448
|
1.706
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr4_-_42658883
|
1.680
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr3_+_194406614
|
1.662
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr19_+_32897021
|
1.651
|
NM_207325
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr16_+_14164879
|
1.646
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr6_+_11538486
|
1.634
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr19_+_32896515
|
1.608
|
NM_001172774
|
DPY19L3
|
dpy-19-like 3 (C. elegans)
|
|
chr8_+_97505876
|
1.604
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr3_-_168865521
|
1.561
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr12_+_3186520
|
1.550
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr12_-_118541655
|
1.544
|
NM_019086
|
VSIG10
|
V-set and immunoglobulin domain containing 10
|
|
chr17_+_25621105
|
1.537
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr14_+_24867926
|
1.535
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr9_-_16870719
|
1.529
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr2_-_44588622
|
1.524
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr3_-_168864325
|
1.474
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_168864066
|
1.457
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr7_+_17338182
|
1.436
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr22_+_45704821
|
1.434
|
NM_001104595
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr6_-_41909386
|
1.422
|
NM_001136125
NM_001760
|
CCND3
|
cyclin D3
|
|
chr2_-_197036301
|
1.419
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr1_-_236228476
|
1.396
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr2_-_44588943
|
1.387
|
NM_001171603
NM_001171617
|
PREPL
|
prolyl endopeptidase-like
|
|
chr16_-_23521803
|
1.343
|
NM_015044
|
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2
|
|
chr6_-_144385647
|
1.336
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr14_+_104605059
|
1.332
|
NM_015656
|
KIF26A
|
kinesin family member 26A
|
|
chr2_-_44586854
|
1.329
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr2_-_44587812
|
1.320
|
NM_006036
|
PREPL
|
prolyl endopeptidase-like
|
|
chr22_+_45705766
|
1.313
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr3_-_32023237
|
1.304
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr12_-_56727720
|
1.298
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr19_-_14316980
|
1.283
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr11_-_89322778
|
1.280
|
NM_001143837
|
NOX4
|
NADPH oxidase 4
|
|
chr16_+_8814567
|
1.261
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr1_-_109935869
|
1.260
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr6_-_42016609
|
1.256
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr22_+_23522551
|
1.249
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr12_-_56727446
|
1.234
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr19_+_10736167
|
1.228
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr16_+_8768403
|
1.225
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr9_-_111882209
|
1.224
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr14_+_63671101
|
1.219
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr1_-_94703252
|
1.198
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr13_+_39612440
|
1.183
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr1_+_25870043
|
1.178
|
NM_015627
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1
|
|
chr14_-_89883306
|
1.174
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr11_-_45687135
|
1.171
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr10_+_129705299
|
1.166
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr1_-_76076762
|
1.151
|
NM_001130058
NM_152697
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr4_+_140586921
|
1.149
|
NM_001204366
NM_001204367
NM_001204368
NM_002413
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr19_+_10713055
|
1.146
|
NM_001145056
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr11_-_89224413
|
1.117
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr3_-_27763784
|
1.116
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr1_+_178694273
|
1.111
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr1_+_76540374
|
1.108
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr6_-_144329342
|
1.104
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chrX_+_150863729
|
1.104
|
NM_024082
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr16_+_8806825
|
1.102
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr8_-_52811707
|
1.102
|
NM_052937
|
PCMTD1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr10_-_101945686
|
1.100
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr21_-_39870303
|
1.068
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr14_-_90085325
|
1.050
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr10_+_129845806
|
1.045
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr21_-_40033617
|
1.045
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr10_-_101945787
|
1.037
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr2_+_109237679
|
1.035
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr12_-_12419702
|
1.021
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr1_+_203595897
|
1.017
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr14_+_66974124
|
1.010
|
NM_001024218
NM_020806
|
GPHN
|
gephyrin
|
|
chrX_-_154033729
|
1.007
|
NM_001166460
NM_001166461
NM_001166462
NM_002436
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa
|
|
chr12_-_31743822
|
1.003
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr3_-_141719188
|
1.003
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr2_+_109150807
|
1.000
|
NM_001193483
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr4_-_16900348
|
0.969
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr1_-_86043932
|
0.968
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr3_+_43328001
|
0.961
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr2_-_158731622
|
0.948
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr11_+_92085261
|
0.942
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr5_-_90679115
|
0.942
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr12_+_56521953
|
0.939
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr2_+_85360373
|
0.930
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr17_+_34136437
|
0.927
|
NM_003487
NM_139215
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa
|
|
chr4_+_108814419
|
0.925
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr2_-_218808705
|
0.916
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr10_-_62703822
|
0.911
|
NM_001242359
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr9_+_91003238
|
0.907
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr2_+_203499819
|
0.898
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr2_+_109223600
|
0.895
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr11_+_134094545
|
0.889
|
NM_052875
|
VPS26B
|
vacuolar protein sorting 26 homolog B (S. pombe)
|
|
chr1_-_109940497
|
0.883
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr11_+_111473099
|
0.879
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chrY_+_16634487
|
0.868
|
NM_001206850
|
NLGN4Y
|
neuroligin 4, Y-linked
|
|
chr14_+_65007163
|
0.851
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr2_+_109271267
|
0.849
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_-_169381417
|
0.844
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_15382874
|
0.831
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr10_+_114709963
|
0.826
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr11_+_842821
|
0.821
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr11_+_844062
|
0.821
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chrX_+_9983793
|
0.818
|
NM_015691
|
WWC3
|
WWC family member 3
|
|
chr16_+_69220940
|
0.815
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr16_-_25026650
|
0.813
|
NM_001006634
NM_018054
|
ARHGAP17
|
Rho GTPase activating protein 17
|
|
chr9_-_74979829
|
0.811
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr17_+_61699786
|
0.811
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr7_+_44646120
|
0.799
|
NM_001003941
NM_001165036
NM_002541
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr2_+_45878912
|
0.797
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr11_+_844445
|
0.795
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr2_+_109204745
|
0.789
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_-_56809594
|
0.786
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr5_+_138677517
|
0.784
|
NM_016480
|
PAIP2
|
poly(A) binding protein interacting protein 2
|
|
chr8_+_86099909
|
0.784
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr1_-_8483701
|
0.783
|
NM_001042682
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr11_-_17191287
|
0.777
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr11_+_111848008
|
0.776
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr2_-_216300770
|
0.775
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr2_-_55646946
|
0.774
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr2_-_158732341
|
0.761
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr16_-_77468930
|
0.760
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr9_-_124984018
|
0.754
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr10_+_97803064
|
0.753
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr6_-_91006580
|
0.752
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr10_+_35535952
|
0.740
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr14_+_103801160
|
0.738
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr9_+_4490193
|
0.738
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr3_-_57113292
|
0.737
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr19_+_54371125
|
0.730
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_+_201798281
|
0.729
|
NM_018085
|
IPO9
|
importin 9
|
|
chr1_+_28764660
|
0.715
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr2_+_173686314
|
0.712
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr7_+_142986680
|
0.707
|
NM_001224
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr8_+_86089618
|
0.703
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr3_-_141868208
|
0.703
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr7_+_142985307
|
0.698
|
NM_032982
NM_032983
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr12_-_109125289
|
0.697
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr5_-_107006585
|
0.696
|
NM_001962
|
EFNA5
|
ephrin-A5
|
|
chr20_-_50384899
|
0.692
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr11_+_121163501
|
0.686
|
NM_001024956
|
SC5DL
|
sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like
|
|
chr17_+_66508098
|
0.677
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr7_-_37024664
|
0.676
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr3_-_15374082
|
0.676
|
NM_004844
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr4_+_108745718
|
0.672
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_-_56835829
|
0.666
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr22_-_36903086
|
0.663
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr22_-_19109912
|
0.659
|
NM_001173533
NM_001173534
NM_001184781
NM_005137
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chr7_-_37393271
|
0.659
|
NM_001206482
|
ELMO1
|
engulfment and cell motility 1
|
|
chr18_+_13611664
|
0.659
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr6_+_80340971
|
0.654
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr10_-_65225605
|
0.653
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr1_-_8877379
|
0.653
|
NM_001042681
NM_012102
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr6_-_32157511
|
0.651
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr2_+_121010413
|
0.649
|
NM_002881
|
RALB
|
v-ral simian leukemia viral oncogene homolog B (ras related; GTP binding protein)
|
|
chr15_-_34629044
|
0.640
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr2_-_197791339
|
0.638
|
NM_024989
|
PGAP1
|
post-GPI attachment to proteins 1
|
|
chr2_+_109204904
|
0.636
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_-_141747434
|
0.634
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|