|
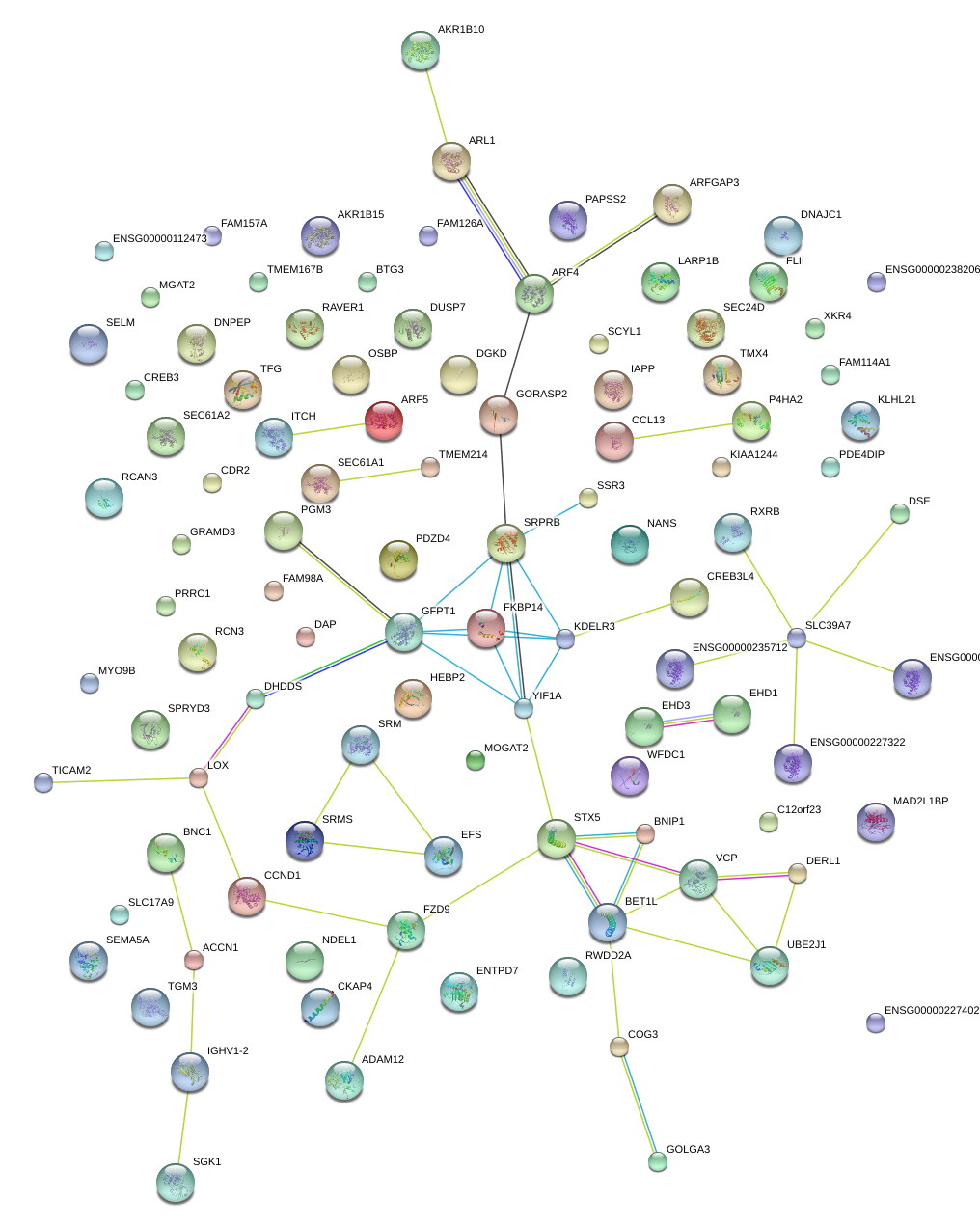
chr5_-_121413906
|
12.691
|
|
LOX
|
lysyl oxidase
|
|
chr5_-_121414011
|
11.191
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr9_+_35732498
|
11.145
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr9_+_35732207
|
10.542
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr9_+_35732629
|
10.424
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr19_+_50031232
|
9.054
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr22_-_31503484
|
6.700
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr2_-_69614321
|
6.610
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr6_-_33168448
|
6.567
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr1_+_26758772
|
6.309
|
NM_001243564
NM_001243565
NM_024887
NM_205861
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr5_-_121413967
|
5.686
|
|
LOX
|
lysyl oxidase
|
|
chr6_+_116692098
|
5.479
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr11_+_69455938
|
5.415
|
|
CCND1
|
cyclin D1
|
|
chr6_-_90062450
|
5.176
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr6_-_90062610
|
5.128
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr11_-_207379
|
5.077
|
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr6_+_83903031
|
5.075
|
NM_033411
|
RWDD2A
|
RWD domain containing 2A
|
|
chr4_+_38869346
|
4.962
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr6_-_83902921
|
4.948
|
NM_001199918
NM_001199919
NM_015599
|
PGM3
|
phosphoglucomutase 3
|
|
chr7_-_30066124
|
4.822
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr11_-_207403
|
4.756
|
NM_001098787
NM_016526
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr11_-_207391
|
4.717
|
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr11_+_65292576
|
4.523
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr16_-_75529278
|
4.377
|
|
|
|
|
chr11_-_62599518
|
4.280
|
NM_001244666
NM_003164
|
STX5
|
syntaxin 5
|
|
chr17_+_8339135
|
4.184
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chr19_+_50030874
|
4.179
|
NM_020650
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr6_-_90062439
|
4.136
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr4_-_119757183
|
4.134
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr11_-_62599447
|
4.025
|
|
STX5
|
syntaxin 5
|
|
chr4_+_128982483
|
3.931
|
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr5_+_126853308
|
3.875
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr19_-_10444186
|
3.840
|
NM_133452
|
RAVER1
|
ribonucleoprotein, PTB-binding 1
|
|
chr1_-_144931967
|
3.825
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr16_-_75529289
|
3.763
|
|
|
|
|
chr6_-_90062349
|
3.760
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr21_-_18985104
|
3.756
|
|
BTG3
|
BTG family, member 3
|
|
chr7_-_23053732
|
3.620
|
NM_032581
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr9_+_100818941
|
3.617
|
NM_018946
|
NANS
|
N-acetylneuraminic acid synthase
|
|
chr1_-_11120001
|
3.577
|
|
SRM
|
spermidine synthase
|
|
chr14_+_50087400
|
3.498
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr3_-_57583080
|
3.497
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr12_-_106641675
|
3.440
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr2_+_171785624
|
3.418
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr13_+_46039024
|
3.346
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr1_-_11120047
|
3.263
|
|
SRM
|
spermidine synthase
|
|
chr19_-_47104456
|
3.243
|
NM_001205281
|
LOC100506012
|
serine/threonine-protein phosphatase 5-like
|
|
chr1_-_11120081
|
3.190
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr4_-_119757288
|
3.172
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr10_+_101419169
|
3.127
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr6_-_134496833
|
3.115
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_+_171785780
|
3.105
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr11_-_59383179
|
3.100
|
|
OSBP
|
oxysterol binding protein
|
|
chr10_+_89419352
|
3.065
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr3_-_52090090
|
3.035
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr16_+_84328400
|
3.014
|
NM_021197
|
WFDC1
|
WAP four-disulfide core domain 1
|
|
chr7_-_23053674
|
2.999
|
|
FAM126A
|
family with sequence similarity 126, member A
|
|
chr3_+_127771342
|
2.981
|
|
|
|
|
chr3_+_127771287
|
2.972
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr12_+_107349673
|
2.959
|
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr6_-_90062289
|
2.955
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr5_-_9546157
|
2.951
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr21_-_18985189
|
2.929
|
|
BTG3
|
BTG family, member 3
|
|
chr22_-_43253186
|
2.908
|
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr6_+_43597278
|
2.897
|
NM_001003690
|
MAD2L1BP
|
MAD2L1 binding protein
|
|
chr3_+_127771307
|
2.872
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr21_-_18985224
|
2.865
|
NM_001130914
NM_006806
|
BTG3
|
BTG family, member 3
|
|
chr1_-_11120076
|
2.847
|
|
SRM
|
spermidine synthase
|
|
chr11_-_64646053
|
2.807
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr22_+_38864015
|
2.746
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr5_-_131563481
|
2.731
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr2_-_33824294
|
2.707
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr2_+_27255745
|
2.705
|
NM_001083590
NM_017727
|
TMEM214
|
transmembrane protein 214
|
|
chr5_-_114938060
|
2.698
|
NM_021649
|
TICAM2
|
toll-like receptor adaptor molecule 2
|
|
chr20_-_8000370
|
2.691
|
NM_021156
|
TMX4
|
thioredoxin-related transmembrane protein 4
|
|
chr3_+_100428393
|
2.645
|
|
TFG
|
TRK-fused gene
|
|
chr6_+_33168602
|
2.615
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr11_-_59383167
|
2.604
|
|
OSBP
|
oxysterol binding protein
|
|
chr1_-_6662681
|
2.542
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr11_-_62598552
|
2.541
|
|
STX5
|
syntaxin 5
|
|
chr19_+_17186569
|
2.538
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr5_+_172571444
|
2.535
|
NM_001205
NM_013978
NM_013979
NM_013980
|
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1
|
|
chr10_-_22292362
|
2.489
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr2_+_27255837
|
2.472
|
|
TMEM214
|
transmembrane protein 214
|
|
chr9_-_35072643
|
2.458
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr1_+_109633373
|
2.454
|
NM_020141
|
TMEM167B
|
transmembrane protein 167B
|
|
chr3_-_57582866
|
2.435
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr7_+_72848108
|
2.414
|
NM_003508
|
FZD9
|
frizzled family receptor 9
|
|
chr20_+_61584094
|
2.406
|
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr15_-_83953465
|
2.377
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr5_-_10761341
|
2.372
|
|
DAP
|
death-associated protein
|
|
chr3_+_100428133
|
2.352
|
NM_001007565
NM_001195479
NM_006070
|
TFG
|
TRK-fused gene
|
|
chr5_-_10761344
|
2.316
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr11_-_59383616
|
2.296
|
NM_002556
|
OSBP
|
oxysterol binding protein
|
|
chr6_+_138483024
|
2.170
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr16_-_22385596
|
2.164
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr11_+_65292526
|
2.149
|
NM_001048218
NM_020680
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr14_-_23834417
|
2.143
|
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr22_-_43253304
|
2.131
|
NM_001142293
NM_014570
|
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3
|
|
chr2_+_234263153
|
2.127
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr8_+_56015013
|
2.111
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr2_+_234263141
|
2.085
|
NM_152879
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr20_+_32951054
|
2.057
|
NM_031483
|
ITCH
|
itchy E3 ubiquitin protein ligase homolog (mouse)
|
|
chr10_-_128077062
|
2.046
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr3_-_156272909
|
2.011
|
NM_007107
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma)
|
|
chr3_+_133524671
|
1.997
|
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr12_-_133405197
|
1.996
|
NM_005895
|
GOLGA3
|
golgin A3
|
|
chr11_-_66056607
|
1.988
|
NM_020470
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr20_-_8000254
|
1.954
|
|
TMX4
|
thioredoxin-related transmembrane protein 4
|
|
chr3_+_133524636
|
1.953
|
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr8_-_124054540
|
1.942
|
|
DERL1
|
Der1-like domain family, member 1
|
|
chr3_-_57583090
|
1.933
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr8_-_124054591
|
1.930
|
NM_001134671
NM_024295
|
DERL1
|
Der1-like domain family, member 1
|
|
chr5_-_10761179
|
1.927
|
|
DAP
|
death-associated protein
|
|
chr12_-_53473171
|
1.915
|
NM_032840
|
SPRYD3
|
SPRY domain containing 3
|
|
chr9_-_35072596
|
1.912
|
|
VCP
|
valosin containing protein
|
|
chr5_+_125759076
|
1.910
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr2_+_64681270
|
1.907
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr12_-_101801499
|
1.891
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr3_-_119182423
|
1.887
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr14_-_39572278
|
1.859
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr19_+_38810540
|
1.844
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr3_-_57583153
|
1.841
|
NM_001660
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr5_+_176730779
|
1.841
|
|
PRELID1
|
PRELI domain containing 1
|
|
chrX_+_68048792
|
1.836
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr17_+_45000493
|
1.831
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr3_+_100428361
|
1.814
|
NM_001195478
|
TFG
|
TRK-fused gene
|
|
chr10_+_98592015
|
1.765
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr5_+_112196884
|
1.756
|
NM_001204193
NM_001204194
NM_001204196
NM_001204199
NM_003135
|
SRP19
|
signal recognition particle 19kDa
|
|
chr6_-_108279378
|
1.752
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr20_+_61583998
|
1.750
|
NM_022082
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr6_+_30294224
|
1.747
|
|
|
|
|
chr7_-_128045995
|
1.690
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr6_-_108279212
|
1.675
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr5_-_89825292
|
1.671
|
NM_198273
|
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3
|
|
chr6_-_108279409
|
1.665
|
NM_007214
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr11_+_61560401
|
1.655
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr14_-_39572414
|
1.653
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr17_+_45000483
|
1.620
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr6_-_108279366
|
1.615
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr12_-_53473167
|
1.614
|
|
SPRYD3
|
SPRY domain containing 3
|
|
chr14_-_24664758
|
1.582
|
NM_001014842
NM_006405
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr5_-_137089434
|
1.581
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr8_-_124054491
|
1.554
|
|
DERL1
|
Der1-like domain family, member 1
|
|
chr14_+_102430767
|
1.553
|
NM_001376
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr5_+_176730812
|
1.518
|
NM_013237
|
PRELID1
|
PRELI domain containing 1
|
|
chr12_-_53473133
|
1.514
|
|
SPRYD3
|
SPRY domain containing 3
|
|
chr4_-_159644439
|
1.509
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr11_-_118927872
|
1.506
|
NM_006389
NM_001130991
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr3_+_122785855
|
1.503
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr10_-_71930185
|
1.483
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr9_-_101984233
|
1.476
|
NM_033087
|
ALG2
|
asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae)
|
|
chr12_+_58120029
|
1.475
|
|
LOC100130776
|
uncharacterized LOC100130776
|
|
chr17_-_74068583
|
1.473
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr12_+_21654745
|
1.450
|
|
GOLT1B
|
golgi transport 1B
|
|
chr17_-_74068582
|
1.445
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr7_+_128095881
|
1.445
|
NM_001098786
NM_013332
|
HILPDA
|
hypoxia inducible lipid droplet-associated
|
|
chr10_-_22292640
|
1.444
|
NM_022365
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr5_+_112196976
|
1.427
|
|
SRP19
|
signal recognition particle 19kDa
|
|
chr5_-_157285977
|
1.425
|
NM_001195555
NM_001195556
NM_014666
|
CLINT1
|
clathrin interactor 1
|
|
chr5_-_140998560
|
1.425
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr17_-_74068597
|
1.423
|
NM_014230
|
SRP68
|
signal recognition particle 68kDa
|
|
chr3_-_128369525
|
1.422
|
|
RPN1
|
ribophorin I
|
|
chr16_-_11836602
|
1.422
|
NM_015914
|
TXNDC11
|
thioredoxin domain containing 11
|
|
chr11_-_66056455
|
1.411
|
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|
|
chr6_-_169653922
|
1.408
|
|
THBS2
|
thrombospondin 2
|
|
chr17_-_74722666
|
1.390
|
|
JMJD6
|
jumonji domain containing 6
|
|
chr3_-_49761293
|
1.381
|
NM_013334
NM_021971
|
GMPPB
|
GDP-mannose pyrophosphorylase B
|
|
chr11_+_61560352
|
1.359
|
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr4_+_110736636
|
1.346
|
NM_018983
NM_032993
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr2_-_118771663
|
1.341
|
|
CCDC93
|
coiled-coil domain containing 93
|
|
chr14_+_102430897
|
1.338
|
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr1_-_145039631
|
1.321
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr17_-_74068522
|
1.318
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr12_+_124069080
|
1.315
|
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr11_+_32112414
|
1.312
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr15_+_41952641
|
1.310
|
|
MGA
|
MAX gene associated
|
|
chr10_-_128077072
|
1.288
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr11_+_65819803
|
1.277
|
NM_006842
|
SF3B2
|
splicing factor 3b, subunit 2, 145kDa
|
|
chr7_-_6523689
|
1.265
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr11_-_118927851
|
1.248
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr16_-_66959418
|
1.244
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr2_-_118771719
|
1.244
|
NM_019044
|
CCDC93
|
coiled-coil domain containing 93
|
|
chr13_-_95248401
|
1.242
|
|
TGDS
|
TDP-glucose 4,6-dehydratase
|
|
chr14_+_52456192
|
1.241
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr13_+_26042917
|
1.240
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr6_-_43596919
|
1.238
|
NM_019096
|
GTPBP2
|
GTP binding protein 2
|
|
chr2_+_220408742
|
1.225
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr17_-_74068576
|
1.220
|
|
SRP68
|
signal recognition particle 68kDa
|
|
chr13_+_26042965
|
1.213
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr17_+_40440481
|
1.211
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr4_-_83350548
|
1.197
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr13_-_95248482
|
1.190
|
NM_014305
|
TGDS
|
TDP-glucose 4,6-dehydratase
|
|
chr5_-_121412805
|
1.175
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr13_+_26828238
|
1.175
|
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr1_+_155108276
|
1.173
|
NM_001122837
NM_001122839
NM_018845
|
SLC50A1
|
solute carrier family 50 (sugar transporter), member 1
|
|
chr4_-_83350829
|
1.156
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr12_+_124069099
|
1.149
|
|
TMED2
|
transmembrane emp24 domain trafficking protein 2
|
|
chr17_+_64298844
|
1.147
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr4_+_76649806
|
1.138
|
|
USO1
|
USO1 vesicle docking protein homolog (yeast)
|