|
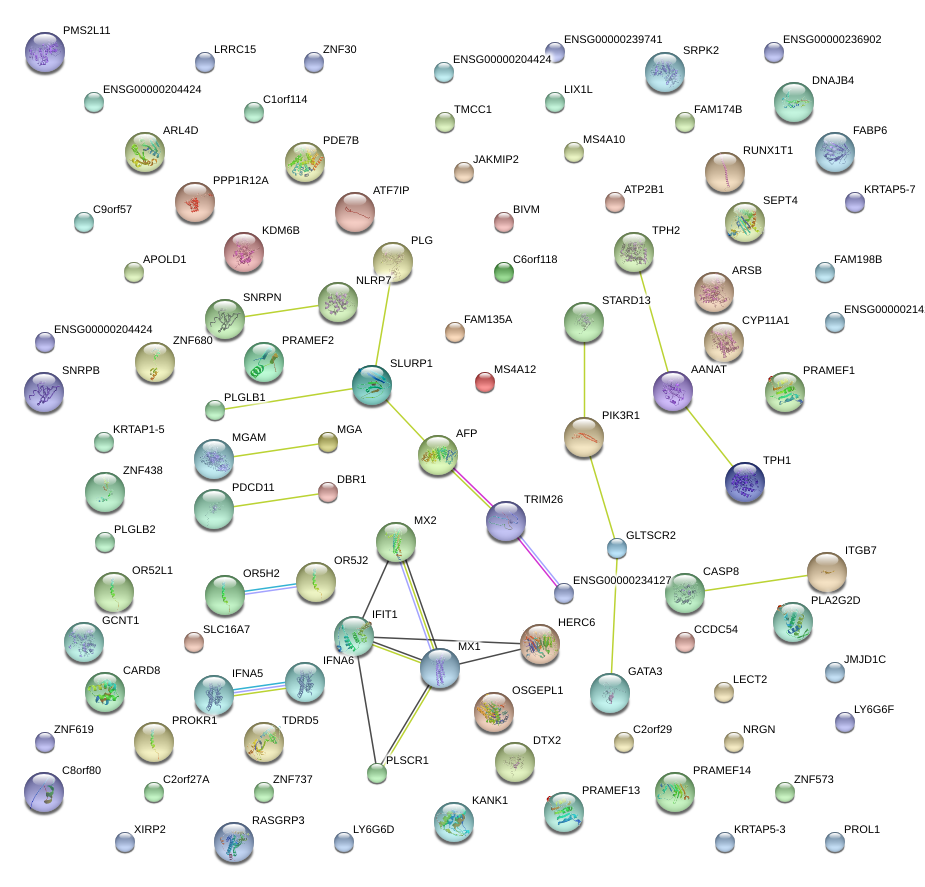
chr6_+_136172802
|
2.886
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr4_+_74301932
|
2.706
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr19_-_48752921
|
2.475
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr19_+_20959109
|
2.428
|
|
|
|
|
chr9_-_74675498
|
2.280
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr19_+_35417806
|
1.941
|
NM_001099437
NM_001099438
NM_194325
|
ZNF30
|
zinc finger protein 30
|
|
chr2_+_202125222
|
1.653
|
NM_033356
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase
|
|
chr19_-_48759141
|
1.414
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr10_-_65028825
|
1.402
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr12_+_14538232
|
1.400
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr5_-_147162264
|
1.394
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_+_78470593
|
1.378
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr12_-_80307690
|
1.348
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr10_+_8097350
|
1.295
|
|
GATA3
|
GATA binding protein 3
|
|
chr8_-_27941387
|
1.271
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr17_-_39183453
|
1.202
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr12_-_53601051
|
1.184
|
|
ITGB7
|
integrin, beta 7
|
|
chr11_+_65266642
|
1.169
|
|
|
|
|
chr15_-_74659926
|
1.149
|
NM_000781
|
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1
|
|
chr19_+_48758931
|
1.136
|
|
LOC100505812
|
uncharacterized LOC100505812
|
|
chr11_+_55944093
|
1.120
|
NM_001005492
|
OR5J2
|
olfactory receptor, family 5, subfamily J, member 2
|
|
chr7_-_64023444
|
1.117
|
|
|
|
|
chr13_-_33780142
|
1.095
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr8_-_93029864
|
1.054
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr19_-_55458864
|
1.043
|
NM_001127255
NM_139176
NM_206828
|
NLRP7
|
NLR family, pyrin domain containing 7
|
|
chr11_-_18062304
|
1.033
|
NM_004179
|
TPH1
|
tryptophan hydroxylase 1
|
|
chr5_+_159656436
|
1.020
|
NM_001445
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr1_-_20445991
|
0.995
|
NM_012400
|
PLA2G2D
|
phospholipase A2, group IID
|
|
chr9_-_21305311
|
0.993
|
NM_002169
|
IFNA5
|
interferon, alpha 5
|
|
chr2_-_190627923
|
0.992
|
NM_022353
|
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1
|
|
chr11_+_65266980
|
0.970
|
|
|
|
|
chr2_+_88047603
|
0.954
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr9_-_3223007
|
0.951
|
|
|
|
|
chr9_+_706744
|
0.948
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr1_+_179575361
|
0.936
|
NM_001199092
|
TDRD5
|
tudor domain containing 5
|
|
chr5_-_78282356
|
0.922
|
NM_000046
|
ARSB
|
arylsulfatase B
|
|
chr10_+_91152321
|
0.919
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr2_-_87248942
|
0.905
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr12_+_12938540
|
0.896
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr15_-_93277303
|
0.895
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr6_+_71136141
|
0.877
|
NM_001162529
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr11_-_1629614
|
0.871
|
NM_001012708
|
KRTAP5-3
|
keratin associated protein 5-3
|
|
chr12_-_90049818
|
0.863
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr4_+_89299874
|
0.861
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr2_+_68872953
|
0.853
|
NM_138964
|
PROKR1
|
prokineticin receptor 1
|
|
chr15_+_42052574
|
0.845
|
|
MGA
|
MAX gene associated
|
|
chr6_-_165723074
|
0.837
|
NM_144980
|
C6orf118
|
chromosome 6 open reading frame 118
|
|
chr11_+_71238312
|
0.822
|
NM_001012503
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr3_+_107096187
|
0.814
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr12_-_46357949
|
0.809
|
|
|
|
|
chr3_-_129599220
|
0.806
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr11_+_60552820
|
0.777
|
NM_206893
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10
|
|
chr19_-_21646685
|
0.772
|
|
|
|
|
chr9_-_21350885
|
0.764
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr3_-_194090459
|
0.759
|
NM_001135057
NM_130830
|
LRRC15
|
leucine rich repeat containing 15
|
|
chr5_+_67588395
|
0.742
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr3_+_40518601
|
0.734
|
NM_001145082
NM_001145093
NM_001145083
|
ZNF619
|
zinc finger protein 619
|
|
chr13_+_103459495
|
0.733
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr7_-_104909442
|
0.724
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr15_+_25281105
|
0.722
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr1_-_13673510
|
0.716
|
NM_001099854
|
PRAMEF14
|
PRAME family member 14
|
|
chr12_+_80603232
|
0.706
|
NM_173591
|
OTOGL
|
otogelin-like
|
|
chr4_-_159094163
|
0.703
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr10_-_31288420
|
0.693
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr10_+_8096772
|
0.688
|
|
GATA3
|
GATA binding protein 3
|
|
chr10_+_8096633
|
0.680
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr12_+_60083125
|
0.680
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr4_+_71263598
|
0.680
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr17_+_74463629
|
0.674
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr7_+_76090971
|
0.668
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr6_+_31683084
|
0.661
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr2_+_132480063
|
0.659
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr5_-_135290521
|
0.658
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr7_-_64023331
|
0.654
|
|
ZNF680
|
zinc finger protein 680
|
|
chr12_-_53600999
|
0.652
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr21_+_42797977
|
0.643
|
NM_002462
NM_001178046
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse)
|
|
chr9_+_79074067
|
0.634
|
NM_001097633
NM_001490
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr2_+_33661392
|
0.632
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr2_+_168043792
|
0.628
|
NM_001199144
NM_001199145
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr3_-_146262421
|
0.625
|
NM_021105
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr19_-_20748540
|
0.622
|
NM_001159293
|
ZNF737
|
zinc finger protein 737
|
|
chr3_-_137893745
|
0.621
|
NM_016216
|
DBR1
|
debranching enzyme homolog 1 (S. cerevisiae)
|
|
chr1_+_145487301
|
0.619
|
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr10_-_27057776
|
0.617
|
|
ABI1
|
abl-interactor 1
|
|
chr10_-_11574273
|
0.615
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr7_+_26400503
|
0.614
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr5_-_997408
|
0.610
|
NM_001242737
|
LOC100506688
|
uncharacterized LOC100506688
|
|
chr4_-_166033924
|
0.604
|
|
TMEM192
|
transmembrane protein 192
|
|
chr2_-_73520448
|
0.604
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr5_+_67586466
|
0.604
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr9_-_113100076
|
0.597
|
NM_001003936
|
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa)
|
|
chr19_-_21950322
|
0.596
|
NM_173531
|
ZNF100
|
zinc finger protein 100
|
|
chr13_-_33859835
|
0.592
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr19_+_37717365
|
0.581
|
NM_152604
|
ZNF383
|
zinc finger protein 383
|
|
chr6_+_147527102
|
0.577
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr3_-_137893718
|
0.575
|
|
DBR1
|
debranching enzyme homolog 1 (S. cerevisiae)
|
|
chrX_-_49089770
|
0.574
|
NM_005183
|
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit
|
|
chr10_-_1071797
|
0.562
|
NM_033261
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2
|
|
chr4_+_106631966
|
0.555
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr9_+_120466453
|
0.545
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr4_-_170077807
|
0.542
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chrX_+_14903412
|
0.539
|
NM_001177475
|
MOSPD2
|
motile sperm domain containing 2
|
|
chr1_+_158979681
|
0.539
|
NM_001206567
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr11_+_5710816
|
0.531
|
NM_001199573
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr12_+_64803741
|
0.530
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr5_-_27038650
|
0.528
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr2_-_47134945
|
0.527
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr19_+_21264952
|
0.524
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr9_+_105757592
|
0.523
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr4_-_159956332
|
0.514
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr1_+_151009028
|
0.514
|
NM_001159642
NM_138278
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like
|
|
chr12_+_15699277
|
0.510
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr8_-_110693233
|
0.508
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr10_+_111765725
|
0.502
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr5_-_88199868
|
0.495
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr10_-_69752385
|
0.492
|
|
HERC4
|
hect domain and RLD 4
|
|
chr4_+_86699858
|
0.492
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_+_179132676
|
0.491
|
|
CANX
|
calnexin
|
|
chrX_-_33146543
|
0.490
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr2_-_45795104
|
0.490
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr2_-_198540583
|
0.488
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr4_+_81256873
|
0.485
|
NM_001206997
NM_152770
|
C4orf22
|
chromosome 4 open reading frame 22
|
|
chr12_-_75905412
|
0.484
|
NM_007043
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr5_+_148731226
|
0.477
|
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr5_-_75008245
|
0.475
|
NM_152408
|
POC5
|
POC5 centriolar protein homolog (Chlamydomonas)
|
|
chr19_-_23869939
|
0.474
|
NM_138330
|
ZNF675
|
zinc finger protein 675
|
|
chr17_+_42634598
|
0.474
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chr1_-_13452655
|
0.472
|
NM_001024661
NM_001099854
|
PRAMEF13
PRAMEF14
|
PRAME family member 13
PRAME family member 14
|
|
chr19_+_52848681
|
0.470
|
NM_173530
|
ZNF610
|
zinc finger protein 610
|
|
chr2_+_109237679
|
0.469
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr1_-_33647626
|
0.468
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chr20_-_45927496
|
0.468
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr6_+_25962978
|
0.466
|
NM_006355
|
TRIM38
|
tripartite motif containing 38
|
|
chr10_-_98325100
|
0.466
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr19_-_14889347
|
0.466
|
NM_013447
NM_152916
NM_152917
NM_152918
NM_152919
NM_152920
NM_152921
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr6_+_79577188
|
0.455
|
NM_001010844
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1
|
|
chr15_+_66585632
|
0.454
|
NM_133375
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like
|
|
chr2_-_61245363
|
0.453
|
NM_144709
|
PUS10
|
pseudouridylate synthase 10
|
|
chr8_-_8175860
|
0.451
|
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr13_+_78315294
|
0.448
|
NM_001242869
NM_001242870
NM_001242871
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr4_-_166034003
|
0.447
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr19_-_20607761
|
0.447
|
|
ZNF826P
|
zinc finger protein 826, pseudogene
|
|
chr19_+_21324826
|
0.446
|
NM_133473
|
ZNF431
|
zinc finger protein 431
|
|
chr4_+_187148624
|
0.446
|
NM_000892
|
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1
|
|
chr11_+_43380403
|
0.444
|
NM_018259
|
TTC17
|
tetratricopeptide repeat domain 17
|
|
chr5_+_159614373
|
0.444
|
NM_001130958
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr1_+_109255555
|
0.444
|
NM_001144937
|
FNDC7
|
fibronectin type III domain containing 7
|
|
chr5_+_126859157
|
0.443
|
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr6_-_51952368
|
0.440
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr1_+_114496498
|
0.437
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr12_+_95676175
|
0.436
|
|
VEZT
|
vezatin, adherens junctions transmembrane protein
|
|
chr15_-_99789585
|
0.435
|
NM_001040655
NM_001040656
NM_001040657
NM_001040658
NM_001040659
NM_001040660
NM_022905
|
TTC23
|
tetratricopeptide repeat domain 23
|
|
chr19_-_53394598
|
0.434
|
NM_207333
|
ZNF320
|
zinc finger protein 320
|
|
chr10_-_99052429
|
0.432
|
NM_001204300
NM_032900
|
ARHGAP19
|
Rho GTPase activating protein 19
|
|
chr12_-_10978867
|
0.430
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chr6_+_12717036
|
0.429
|
NM_001242648
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr3_-_119963141
|
0.425
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr12_-_15038724
|
0.423
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr12_+_113376001
|
0.421
|
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa
|
|
chr19_-_48614013
|
0.420
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chrX_+_109662284
|
0.420
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr13_+_49684388
|
0.417
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr14_-_45722332
|
0.416
|
|
MIS18BP1
|
MIS18 binding protein 1
|
|
chr5_-_88119604
|
0.415
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_-_173962050
|
0.415
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr12_-_75905376
|
0.412
|
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr15_-_77176191
|
0.408
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr16_-_71610948
|
0.407
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr4_+_175839508
|
0.404
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr18_+_28898051
|
0.404
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr4_+_186990308
|
0.402
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr17_-_54893249
|
0.401
|
NM_001085430
|
C17orf67
|
chromosome 17 open reading frame 67
|
|
chr4_-_122791468
|
0.400
|
NM_018190
NM_176824
|
BBS7
|
Bardet-Biedl syndrome 7
|
|
chr12_+_7942025
|
0.398
|
|
NANOG
|
Nanog homeobox
|
|
chr1_+_210502188
|
0.396
|
NM_001170587
NM_001170588
NM_018194
|
HHAT
|
hedgehog acyltransferase
|
|
chr7_-_64023454
|
0.394
|
NM_001130022
NM_178558
|
ZNF680
|
zinc finger protein 680
|
|
chr10_-_99052391
|
0.394
|
|
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough
Rho GTPase activating protein 19
|
|
chr19_-_50666279
|
0.393
|
NM_152358
|
IZUMO2
|
IZUMO family member 2
|
|
chr11_-_62457001
|
0.392
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr5_-_149466173
|
0.391
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr1_-_85742586
|
0.386
|
NM_003921
|
BCL10
|
B-cell CLL/lymphoma 10
|
|
chr16_-_11375094
|
0.386
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr11_-_107436423
|
0.384
|
NM_138775
|
ALKBH8
|
alkB, alkylation repair homolog 8 (E. coli)
|
|
chr20_-_1538303
|
0.383
|
NM_178460
|
SIRPD
|
signal-regulatory protein delta
|
|
chr3_-_137893636
|
0.383
|
|
DBR1
|
debranching enzyme homolog 1 (S. cerevisiae)
|
|
chr15_-_74658498
|
0.380
|
NM_001099773
|
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1
|
|
chr11_+_5641173
|
0.379
|
NM_001003827
|
TRIM34
|
tripartite motif containing 34
|
|
chr12_-_75905391
|
0.378
|
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr1_+_84768333
|
0.378
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr5_+_102465256
|
0.378
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chrX_+_30261847
|
0.375
|
NM_002363
NM_177415
|
MAGEB1
|
melanoma antigen family B, 1
|
|
chr11_+_33363104
|
0.370
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr8_+_62200524
|
0.370
|
NM_173519
|
CLVS1
|
clavesin 1
|
|
chr3_-_9920633
|
0.369
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr1_+_207262211
|
0.367
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr5_+_147774274
|
0.362
|
|
FBXO38
|
F-box protein 38
|
|
chr2_+_168675181
|
0.361
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr3_+_130279177
|
0.357
|
NM_001102608
|
COL6A6
|
collagen, type VI, alpha 6
|
|
chr21_-_35884503
|
0.356
|
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr13_+_42712177
|
0.355
|
NM_001204505
NM_001204506
|
DGKH
|
diacylglycerol kinase, eta
|