|
chr21_+_46293925
|
3.500
|
|
|
|
|
chr17_-_31204071
|
3.038
|
|
MYO1D
|
myosin ID
|
|
chr7_-_22396511
|
2.919
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr19_+_10381763
|
2.250
|
|
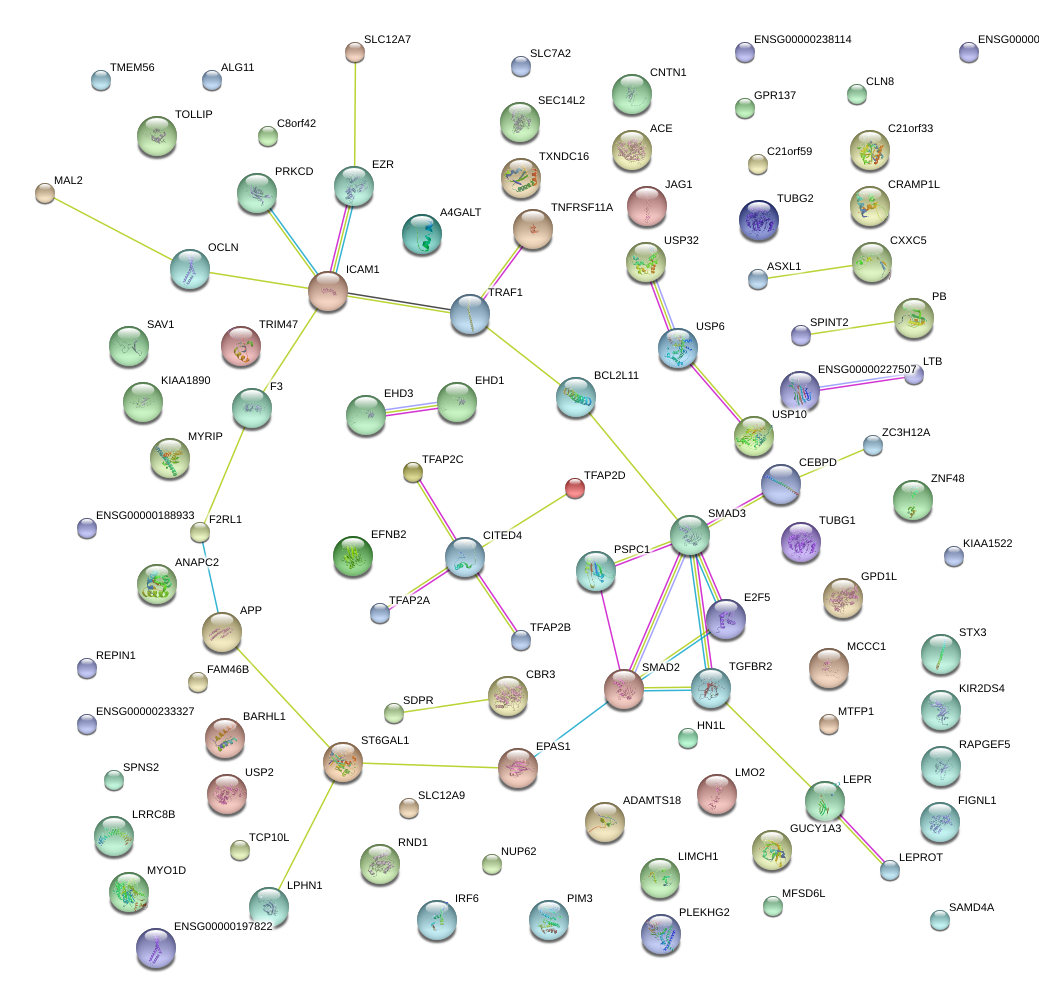
ICAM1
|
intercellular adhesion molecule 1
|
|
chr19_+_38755238
|
2.182
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_38755097
|
2.159
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_38755422
|
2.081
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr15_+_67358194
|
1.721
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr1_-_209979388
|
1.622
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr14_-_51134862
|
1.438
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr14_-_51135021
|
1.431
|
NM_021818
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr1_-_95007139
|
1.413
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr1_+_37940180
|
1.338
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr19_-_50432731
|
1.324
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr1_+_37940129
|
1.276
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr1_+_37940074
|
1.229
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr16_+_84733606
|
1.218
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr18_+_59992519
|
1.200
|
NM_003839
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator
|
|
chr3_+_30648372
|
1.168
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr5_+_139028483
|
1.166
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr2_-_28113954
|
1.119
|
|
LOC100302650
|
uncharacterized LOC100302650
|
|
chr5_+_139027868
|
1.108
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr6_-_10419786
|
1.081
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr5_+_139027907
|
1.080
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr19_+_10381516
|
1.073
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr5_-_1112106
|
1.067
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr22_-_43089856
|
1.047
|
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr1_+_65991390
|
1.046
|
|
LEPR
|
leptin receptor
|
|
chr5_+_139027955
|
1.041
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr1_-_27339448
|
1.034
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr5_+_76114881
|
1.016
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr3_+_39851028
|
0.995
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr13_-_107187312
|
0.991
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr4_+_156588811
|
0.989
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr8_+_17354562
|
0.980
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr3_-_182833859
|
0.955
|
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha)
|
|
chr21_+_26934440
|
0.954
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr7_-_50518021
|
0.944
|
NM_001042762
NM_022116
|
FIGNL1
|
fidgetin-like 1
|
|
chr14_-_61748193
|
0.928
|
|
|
|
|
chr6_-_159240398
|
0.928
|
|
EZR
|
ezrin
|
|
chr17_-_73874056
|
0.920
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr2_+_46524901
|
0.917
|
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr4_+_2420671
|
0.907
|
NM_001193282
|
LOC402160
|
uncharacterized LOC402160
|
|
chr8_-_48650683
|
0.862
|
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr5_+_139028025
|
0.859
|
NM_016463
|
CXXC5
|
CXXC finger protein 5
|
|
chr19_+_39903749
|
0.858
|
NM_022835
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2
|
|
chr11_-_2018703
|
0.847
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr17_+_4402128
|
0.846
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr6_-_159240399
|
0.837
|
|
EZR
|
ezrin
|
|
chr7_+_150065330
|
0.829
|
|
REPIN1
|
replication initiator 1
|
|
chr5_+_76114832
|
0.828
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr21_-_33984657
|
0.808
|
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr1_-_41328002
|
0.805
|
NM_133467
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr1_+_89990396
|
0.792
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr21_+_37507209
|
0.789
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr3_+_53195214
|
0.787
|
NM_006254
NM_212539
|
PRKCD
|
protein kinase C, delta
|
|
chr1_-_41328019
|
0.784
|
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4
|
|
chr22_+_50354139
|
0.783
|
NM_001001852
|
PIM3
|
pim-3 oncogene
|
|
chr11_-_119234628
|
0.773
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr2_+_111878450
|
0.771
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr3_+_186648515
|
0.766
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr6_-_31550065
|
0.765
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr19_-_50432678
|
0.758
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr11_-_2018568
|
0.757
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr16_+_30406711
|
0.750
|
NM_152652
|
ZNF48
|
zinc finger protein 48
|
|
chr8_+_86089641
|
0.743
|
|
|
|
|
chr16_+_84733611
|
0.738
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr8_+_86089618
|
0.722
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr3_+_32148000
|
0.713
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr11_-_1330891
|
0.710
|
NM_019009
|
TOLLIP
|
toll interacting protein
|
|
chr21_+_45553485
|
0.710
|
NM_004649
NM_198155
|
C21orf33
|
chromosome 21 open reading frame 33
|
|
chr3_+_32148136
|
0.708
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr11_-_33890931
|
0.704
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr9_-_123691442
|
0.700
|
NM_001190945
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr12_-_49259549
|
0.697
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr14_-_51135041
|
0.693
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr8_+_1712011
|
0.684
|
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr17_+_40811261
|
0.676
|
NM_016437
|
TUBG2
|
tubulin, gamma 2
|
|
chr9_-_140083034
|
0.664
|
NM_013366
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr11_-_1330883
|
0.661
|
|
TOLLIP
|
toll interacting protein
|
|
chr9_+_135457974
|
0.661
|
NM_020064
|
BARHL1
|
BarH-like homeobox 1
|
|
chr7_+_100450327
|
0.656
|
|
SLC12A9
|
solute carrier family 12 (potassium/chloride transporters), member 9
|
|
chr11_-_64646053
|
0.641
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr9_-_140082983
|
0.641
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr4_+_156587861
|
0.639
|
NM_000856
NM_001130682
NM_001130686
NM_001130687
NM_001130683
NM_001130685
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr8_-_48650703
|
0.639
|
NM_005195
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr21_-_33984455
|
0.628
|
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr20_+_30946786
|
0.627
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr11_-_2019070
|
0.624
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr20_-_10654139
|
0.623
|
|
JAG1
|
jagged 1
|
|
chr8_+_120220554
|
0.620
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr1_+_95582827
|
0.609
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr4_+_41362750
|
0.609
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr13_+_52586516
|
0.607
|
NM_001004127
|
ALG11
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
|
|
chr17_+_61554420
|
0.600
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr19_-_18391440
|
0.600
|
|
|
|
|
chr19_-_14316980
|
0.599
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr16_+_84733596
|
0.598
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr5_+_68788387
|
0.596
|
NM_001205254
|
OCLN
|
occludin
|
|
chr16_-_77468930
|
0.587
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr17_-_8702438
|
0.585
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr13_-_20357125
|
0.584
|
NM_001042414
|
PSPC1
|
paraspeckle component 1
|
|
chr2_-_192711651
|
0.583
|
|
SDPR
|
serum deprivation response
|
|
chr21_-_27542919
|
0.577
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr22_+_30792929
|
0.568
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr21_-_33984865
|
0.567
|
NM_021254
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr11_-_33891258
|
0.567
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr20_-_10654429
|
0.566
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr1_+_33207343
|
0.563
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr16_+_1728290
|
0.551
|
|
HN1L
|
hematological and neurological expressed 1-like
|
|
chr11_+_59522905
|
0.548
|
|
STX3
|
syntaxin 3
|
|
chr14_+_55034329
|
0.545
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr6_-_159240367
|
0.539
|
|
EZR
|
ezrin
|
|
chr8_-_494834
|
0.529
|
|
C8orf42
|
chromosome 8 open reading frame 42
|
|
chr1_+_65991357
|
0.526
|
NM_001198687
NM_001198688
NM_001198689
|
LEPR
|
leptin receptor
|
|
chr5_+_31639424
|
0.526
|
|
PDZD2
|
PDZ domain containing 2
|
|
chr11_-_1330840
|
0.525
|
|
TOLLIP
|
toll interacting protein
|
|
chr16_-_11680756
|
0.516
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr1_+_12123433
|
0.514
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr1_-_3528058
|
0.514
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr12_-_121734543
|
0.513
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr6_+_142622797
|
0.513
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr16_+_84733554
|
0.513
|
NM_005153
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr5_-_88199868
|
0.509
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_173600634
|
0.507
|
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr6_-_159240433
|
0.506
|
NM_001111077
|
EZR
|
ezrin
|
|
chr15_-_69113213
|
0.504
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr15_-_69113089
|
0.501
|
|
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A
|
|
chr4_-_151936648
|
0.500
|
NM_006726
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr11_-_1330808
|
0.496
|
|
TOLLIP
|
toll interacting protein
|
|
chr11_-_694842
|
0.495
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr8_-_141467810
|
0.494
|
|
TRAPPC9
|
trafficking protein particle complex 9
|
|
chr1_-_51425795
|
0.491
|
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr2_-_24583291
|
0.490
|
NM_006277
NM_019595
NM_147152
|
ITSN2
|
intersectin 2
|
|
chr6_+_80340971
|
0.489
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr4_-_4291817
|
0.489
|
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr2_-_208634051
|
0.487
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr8_+_37654833
|
0.486
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr4_-_4291731
|
0.485
|
NM_001145725
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr19_+_18391252
|
0.480
|
|
|
|
|
chr5_+_32173481
|
0.479
|
|
LOC153546
|
uncharacterized LOC153546
|
|
chr17_-_43510279
|
0.478
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr22_+_31608284
|
0.478
|
|
LIMK2
|
LIM domain kinase 2
|
|
chr1_-_51425740
|
0.476
|
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr7_-_37488894
|
0.474
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr2_-_235405692
|
0.473
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr13_-_20357081
|
0.473
|
|
PSPC1
|
paraspeckle component 1
|
|
chr15_-_52587851
|
0.473
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr12_-_121734474
|
0.471
|
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr1_-_27339326
|
0.469
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr17_-_31204187
|
0.469
|
|
MYO1D
|
myosin ID
|
|
chr13_+_114238978
|
0.469
|
|
TFDP1
|
transcription factor Dp-1
|
|
chr6_+_144471631
|
0.469
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr22_+_31608145
|
0.468
|
|
LIMK2
|
LIM domain kinase 2
|
|
chr11_-_2018446
|
0.466
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr4_-_4291889
|
0.459
|
NM_017816
|
LYAR
|
Ly1 antibody reactive homolog (mouse)
|
|
chr19_-_2783225
|
0.456
|
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr11_-_71159371
|
0.456
|
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr17_+_32597239
|
0.455
|
NM_006273
|
CCL7
|
chemokine (C-C motif) ligand 7
|
|
chr22_+_41777932
|
0.451
|
NM_003216
|
TEF
|
thyrotrophic embryonic factor
|
|
chr4_+_139936938
|
0.450
|
NM_012118
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae)
|
|
chr2_+_28616346
|
0.450
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr8_-_141467853
|
0.448
|
NM_001160372
|
TRAPPC9
|
trafficking protein particle complex 9
|
|
chr19_+_55591759
|
0.448
|
NM_017729
|
EPS8L1
|
EPS8-like 1
|
|
chr22_+_50354428
|
0.447
|
|
PIM3
|
pim-3 oncogene
|
|
chr1_-_51425918
|
0.446
|
NM_007051
|
FAF1
|
Fas (TNFRSF6) associated factor 1
|
|
chr19_-_2783313
|
0.445
|
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr5_+_56110899
|
0.444
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr4_+_4291923
|
0.440
|
NM_145291
|
ZBTB49
|
zinc finger and BTB domain containing 49
|
|
chr6_+_142623364
|
0.440
|
|
GPR126
|
G protein-coupled receptor 126
|
|
chr14_-_105635114
|
0.437
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr12_+_79439432
|
0.435
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr6_+_53659738
|
0.434
|
NM_018214
|
LRRC1
|
leucine rich repeat containing 1
|
|
chr14_-_55369163
|
0.433
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr1_+_78511588
|
0.431
|
NM_017655
|
GIPC2
|
GIPC PDZ domain containing family, member 2
|
|
chr19_-_7701852
|
0.427
|
|
|
|
|
chr2_+_28616360
|
0.426
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr20_+_61448391
|
0.424
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr13_-_77460432
|
0.422
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr14_-_76127526
|
0.421
|
NM_007176
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr2_+_46524448
|
0.421
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr19_-_50432679
|
0.420
|
|
NUP62
IL4I1
|
nucleoporin 62kDa
interleukin 4 induced 1
|
|
chr22_+_35796294
|
0.419
|
|
MCM5
|
minichromosome maintenance complex component 5
|
|
chr21_+_45553546
|
0.418
|
|
C21orf33
|
chromosome 21 open reading frame 33
|
|
chr14_-_105634708
|
0.416
|
|
JAG2
|
jagged 2
|
|
chr19_-_2783332
|
0.415
|
NM_003021
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr20_+_56725982
|
0.413
|
NM_178456
|
C20orf85
|
chromosome 20 open reading frame 85
|
|
chr12_+_122459791
|
0.413
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr6_+_36646455
|
0.410
|
NM_000389
NM_001220778
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr4_+_129732427
|
0.408
|
|
PHF17
|
PHD finger protein 17
|
|
chr6_-_47277640
|
0.406
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr19_-_1132109
|
0.402
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr11_-_69590170
|
0.401
|
NM_002007
|
FGF4
|
fibroblast growth factor 4
|
|
chr9_+_131185129
|
0.400
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr7_+_150065968
|
0.400
|
|
REPIN1
|
replication initiator 1
|
|
chr2_+_170590454
|
0.400
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr3_-_47517417
|
0.397
|
NM_012235
|
SCAP
|
SREBF chaperone
|
|
chr5_-_32313069
|
0.394
|
NM_001040446
|
MTMR12
|
myotubularin related protein 12
|
|
chr19_+_35739778
|
0.392
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr11_+_65779461
|
0.384
|
NM_001323
|
CST6
|
cystatin E/M
|