|
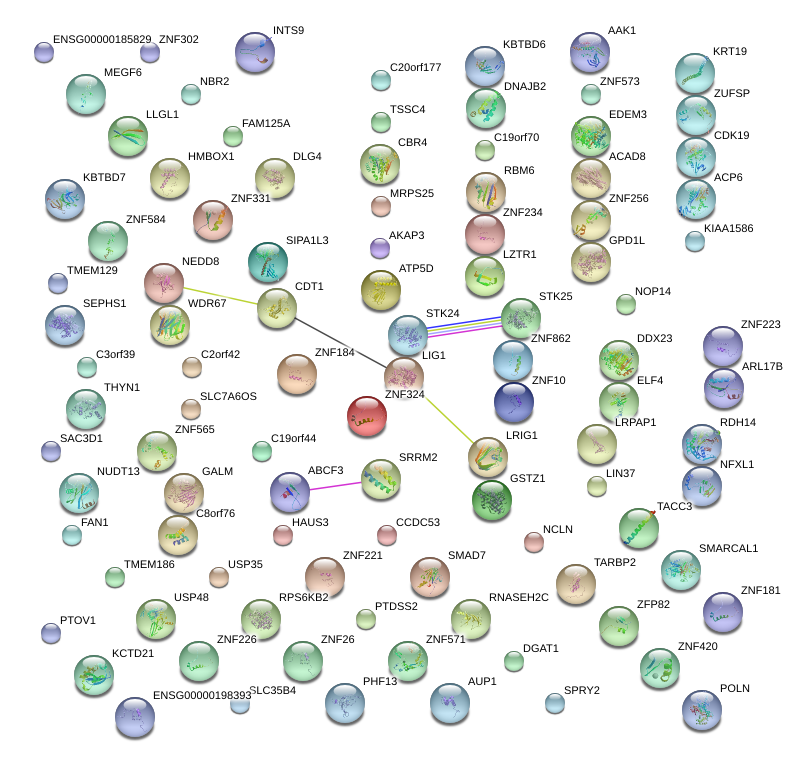
chr19_+_44556163
|
2.193
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr3_+_49977476
|
1.700
|
NM_001167582
NM_005777
|
RBM6
|
RNA binding motif protein 6
|
|
chr3_+_49977582
|
1.666
|
|
RBM6
|
RNA binding motif protein 6
|
|
chr19_+_54024176
|
1.587
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr19_-_48673559
|
1.464
|
NM_000234
|
LIG1
|
ligase I, DNA, ATP-dependent
|
|
chr4_+_1723212
|
1.373
|
NM_006342
|
TACC3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr6_-_27440459
|
1.363
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr8_-_28747697
|
1.361
|
NM_001145159
NM_018250
|
INTS9
|
integrator complex subunit 9
|
|
chr19_+_3185860
|
1.221
|
NM_020170
|
NCLN
|
nicalin
|
|
chr4_-_47916498
|
1.197
|
NM_152995
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chr3_+_32148000
|
1.178
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr19_+_50353942
|
1.167
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr19_+_16607181
|
1.143
|
NM_032207
|
C19orf44
|
chromosome 19 open reading frame 44
|
|
chr11_+_64808375
|
1.119
|
NM_013299
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr8_-_28747422
|
1.104
|
NM_001172562
|
INTS9
|
integrator complex subunit 9
|
|
chr3_-_43147470
|
1.103
|
NM_032806
|
C3orf39
|
chromosome 3 open reading frame 39
|
|
chr11_-_77899640
|
1.096
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr3_-_15106806
|
1.053
|
NM_022497
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr14_+_77787229
|
1.049
|
NM_145870
NM_145871
|
GSTZ1
|
glutathione transferase zeta 1
|
|
chr1_+_6673744
|
1.049
|
NM_153812
|
PHF13
|
PHD finger protein 13
|
|
chr11_-_65488142
|
1.042
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr11_+_64808395
|
1.035
|
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr3_-_15106701
|
1.021
|
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr19_-_36705556
|
1.001
|
NM_152477
|
ZNF565
|
zinc finger protein 565
|
|
chr12_+_53895065
|
0.998
|
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr3_+_32148136
|
0.996
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr18_-_77439267
|
0.991
|
|
|
|
|
chr19_-_5680910
|
0.975
|
NM_205767
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr8_+_28747885
|
0.973
|
NM_001135726
|
HMBOX1
|
homeobox containing 1
|
|
chr19_+_36239445
|
0.960
|
|
LIN37
|
lin-37 homolog (C. elegans)
|
|
chr4_-_1722978
|
0.956
|
NM_001127266
NM_138385
|
TMEM129
|
transmembrane protein 129
|
|
chr11_+_450346
|
0.946
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr13_-_41768533
|
0.927
|
NM_032138
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7
|
|
chr4_+_1723233
|
0.920
|
|
TACC3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr11_+_450260
|
0.917
|
NM_030783
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr19_+_44669214
|
0.906
|
NM_001146220
NM_001032373
NM_001032374
NM_015919
NM_001032372
|
ZNF226
|
zinc finger protein 226
|
|
chr12_+_53895376
|
0.899
|
NM_134324
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr16_+_2802634
|
0.890
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr1_-_184723952
|
0.865
|
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr4_-_169931387
|
0.864
|
NM_032783
|
CBR4
|
carbonyl reductase 4
|
|
chr4_-_2965147
|
0.848
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr19_+_44645728
|
0.848
|
|
ZNF234
|
zinc finger protein 234
|
|
chr11_-_65487910
|
0.847
|
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr19_+_35225095
|
0.846
|
|
ZNF181
|
zinc finger protein 181
|
|
chr8_-_145550536
|
0.844
|
NM_012079
|
DGAT1
|
diacylglycerol O-acyltransferase 1
|
|
chr2_-_74756815
|
0.841
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr11_+_67195886
|
0.838
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr11_+_2421983
|
0.828
|
|
TSSC4
|
tumor suppressing subtransferable candidate 4
|
|
chr2_-_74756838
|
0.826
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr19_+_44331443
|
0.825
|
NM_181845
|
ZNF283
|
zinc finger protein 283
|
|
chr2_-_74756823
|
0.824
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr13_-_41706587
|
0.823
|
NM_152903
|
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6
|
|
chr2_-_70418086
|
0.822
|
NM_017880
|
C2orf42
|
chromosome 2 open reading frame 42
|
|
chr20_+_58515408
|
0.801
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr19_-_58459013
|
0.795
|
NM_005773
|
ZNF256
|
zinc finger protein 256
|
|
chr19_-_36705982
|
0.792
|
NM_001042474
|
ZNF565
|
zinc finger protein 565
|
|
chr19_+_35168558
|
0.785
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr2_-_74756973
|
0.771
|
NM_181575
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr22_+_21336508
|
0.769
|
NM_006767
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr19_-_38270199
|
0.752
|
NM_001172689
NM_001172690
NM_001172691
NM_001172692
NM_152360
|
ZNF573
|
zinc finger protein 573
|
|
chr22_+_21336616
|
0.751
|
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr1_-_147142341
|
0.749
|
NM_016361
|
ACP6
|
acid phosphatase 6, lysophosphatidic
|
|
chr16_+_2802683
|
0.734
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr1_-_22109669
|
0.710
|
NM_001032730
NM_032236
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr2_+_220144024
|
0.706
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr13_-_80915041
|
0.705
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr1_+_156024516
|
0.702
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr17_-_7123368
|
0.695
|
NM_001365
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr12_-_102455763
|
0.694
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr2_-_242447449
|
0.689
|
|
STK25
|
serine/threonine kinase 25
|
|
chr2_-_69870705
|
0.689
|
|
AAK1
|
AP2 associated kinase 1
|
|
chr19_+_38397880
|
0.672
|
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr8_-_124253584
|
0.671
|
NM_032847
|
C8orf76
|
chromosome 8 open reading frame 76
|
|
chr14_+_77787705
|
0.670
|
NM_001513
|
GSTZ1
|
glutathione transferase zeta 1
|
|
chr18_-_46477080
|
0.666
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr3_+_183903970
|
0.665
|
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr2_-_18741837
|
0.657
|
NM_020905
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis)
|
|
chr11_+_77899857
|
0.655
|
NM_020798
|
USP35
|
ubiquitin specific peptidase 35
|
|
chr19_+_3185965
|
0.654
|
|
NCLN
|
nicalin
|
|
chr3_+_183903862
|
0.654
|
NM_018358
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr19_+_58978336
|
0.653
|
NM_014347
|
ZNF324
|
zinc finger protein 324
|
|
chr16_+_2802676
|
0.625
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr2_+_217277136
|
0.619
|
NM_014140
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1
|
|
chr12_-_4758175
|
0.618
|
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr6_-_111136288
|
0.617
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr19_+_1241742
|
0.617
|
NM_001001975
NM_001687
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr14_-_24701561
|
0.616
|
NM_001199823
NM_006156
|
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr11_+_134123406
|
0.614
|
NM_014384
|
ACAD8
|
acyl-CoA dehydrogenase family, member 8
|
|
chr4_-_3534138
|
0.612
|
|
LRPAP1
|
low density lipoprotein receptor-related protein associated protein 1
|
|
chr11_-_134123138
|
0.606
|
NM_001037304
NM_001037305
NM_014174
NM_199297
NM_199298
|
THYN1
|
thymocyte nuclear protein 1
|
|
chr19_+_50354368
|
0.606
|
NM_017432
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr19_-_36909463
|
0.603
|
NM_133466
|
ZFP82
|
zinc finger protein 82 homolog (mouse)
|
|
chr7_-_134001749
|
0.601
|
NM_032826
|
SLC35B4
|
solute carrier family 35, member B4
|
|
chr1_-_3447974
|
0.600
|
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr6_-_116989901
|
0.599
|
NM_145062
|
ZUFSP
|
zinc finger with UFM1-specific peptidase domain
|
|
chr4_-_2243847
|
0.597
|
NM_024511
|
POLN
HAUS3
|
polymerase (DNA directed) nu
HAUS augmin-like complex, subunit 3
|
|
chr16_+_2802640
|
0.596
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr10_+_74870209
|
0.595
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chr16_+_2802652
|
0.594
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr16_-_68344785
|
0.593
|
NM_032178
|
SLC7A6OS
|
solute carrier family 7, member 6 opposite strand
|
|
chr16_-_8891472
|
0.591
|
NM_015421
|
TMEM186
|
transmembrane protein 186
|
|
chr19_+_1241811
|
0.590
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr12_+_133757994
|
0.583
|
NM_001165883
NM_001165884
NM_001165885
NM_001165886
NM_001165887
NM_003415
NM_152943
NM_001165881
NM_001165882
|
ZNF268
|
zinc finger protein 268
|
|
chr16_+_2802319
|
0.580
|
NM_016333
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr15_+_31196106
|
0.577
|
|
FAN1
|
FANCD2/FANCI-associated nuclease 1
|
|
chr10_-_13390189
|
0.575
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr10_-_43048029
|
0.574
|
|
ZNF37BP
|
zinc finger protein 37B, pseudogene
|
|
chr19_+_37569307
|
0.573
|
NM_144689
|
ZNF420
|
zinc finger protein 420
|
|
chr19_+_35225479
|
0.571
|
NM_001029997
NM_001145665
|
ZNF181
|
zinc finger protein 181
|
|
chr8_+_124084898
|
0.565
|
NM_001145088
NM_145647
|
WDR67
|
WD repeat domain 67
|
|
chr12_-_49245904
|
0.564
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr19_+_1241766
|
0.563
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr19_+_17530841
|
0.560
|
|
FAM125A
|
family with sequence similarity 125, member A
|
|
chr7_+_149535478
|
0.559
|
|
ZNF862
|
zinc finger protein 862
|
|
chr17_-_39684585
|
0.554
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr12_+_133562941
|
0.553
|
NM_019591
|
ZNF26
|
zinc finger protein 26
|
|
chr6_+_56911310
|
0.550
|
NM_020931
|
KIAA1586
|
KIAA1586
|
|
chr19_+_1241767
|
0.550
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr17_+_41277608
|
0.545
|
|
NBR2
|
neighbor of BRCA1 gene 2 (non-protein coding)
|
|
chr2_+_38893051
|
0.542
|
NM_138801
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr11_-_69634191
|
0.542
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chr13_-_53313835
|
0.540
|
|
LECT1
|
leukocyte cell derived chemotaxin 1
|
|
chr16_-_3285359
|
0.537
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr19_-_44617271
|
0.537
|
|
LOC100379224
|
uncharacterized LOC100379224
|
|
chr20_+_18269120
|
0.534
|
NM_001083330
NM_003434
|
ZNF133
|
zinc finger protein 133
|
|
chr6_+_42531736
|
0.533
|
NM_001184801
NM_015255
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr19_-_44809126
|
0.530
|
NM_004234
|
ZNF235
|
zinc finger protein 235
|
|
chr19_-_5680503
|
0.528
|
|
C19orf70
|
chromosome 19 open reading frame 70
|
|
chr12_+_53894704
|
0.527
|
NM_004178
NM_134323
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr19_+_3185927
|
0.523
|
|
NCLN
|
nicalin
|
|
chr19_-_38085646
|
0.522
|
NM_016536
|
ZNF571
|
zinc finger protein 571
|
|
chr16_+_2802673
|
0.522
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr19_-_44860798
|
0.516
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr2_-_190445612
|
0.514
|
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr19_+_507475
|
0.513
|
NM_033513
|
TPGS1
|
tubulin polyglutamylase complex subunit 1
|
|
chr19_-_54619025
|
0.513
|
NM_013342
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr2_-_74756755
|
0.512
|
|
AUP1
|
ancient ubiquitous protein 1
|
|
chr16_-_30934437
|
0.511
|
|
FBXL19-AS1
|
FBXL19 antisense RNA 1 (non-protein coding)
|
|
chr19_+_58790306
|
0.507
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr6_+_56911422
|
0.499
|
|
KIAA1586
|
KIAA1586
|
|
chr19_+_44529493
|
0.497
|
NM_001129996
NM_013360
|
ZNF284
ZNF222
|
zinc finger protein 284
zinc finger protein 222
|
|
chr19_-_44439410
|
0.494
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chr6_+_15245702
|
0.493
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr19_+_50354417
|
0.486
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr9_-_138853146
|
0.484
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr14_-_24701559
|
0.483
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr1_-_185126153
|
0.483
|
NM_001202423
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr2_+_217277422
|
0.481
|
NM_001127207
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1
|
|
chr19_+_58978637
|
0.478
|
|
ZNF324
|
zinc finger protein 324
|
|
chr1_-_41131113
|
0.476
|
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr7_+_76090971
|
0.476
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr20_-_32891075
|
0.474
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr12_-_2986095
|
0.474
|
|
FOXM1
|
forkhead box M1
|
|
chr16_-_3285069
|
0.473
|
NM_001145447
NM_003454
|
ZNF200
|
zinc finger protein 200
|
|
chr3_-_186524226
|
0.473
|
NM_002916
|
RFC4
|
replication factor C (activator 1) 4, 37kDa
|
|
chr19_+_17530856
|
0.473
|
NM_138401
|
FAM125A
|
family with sequence similarity 125, member A
|
|
chr6_+_56954889
|
0.472
|
|
ZNF451
|
zinc finger protein 451
|
|
chr5_+_178450775
|
0.470
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr19_-_49222956
|
0.468
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr1_-_155532322
|
0.467
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr16_-_4784153
|
0.466
|
NM_133450
NM_001242929
|
ANKS3
|
ankyrin repeat and sterile alpha motif domain containing 3
|
|
chr17_-_39684499
|
0.466
|
|
KRT19
|
keratin 19
|
|
chr12_-_133532847
|
0.465
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr11_-_66206260
|
0.463
|
NM_016050
NM_170738
NM_170739
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr8_+_99129488
|
0.463
|
NM_001145860
NM_001145861
|
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae)
|
|
chr16_-_4588379
|
0.459
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr16_+_68344868
|
0.458
|
NM_001184824
NM_019023
|
PRMT7
|
protein arginine methyltransferase 7
|
|
chrX_-_152938742
|
0.457
|
NM_001135740
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr22_+_18560623
|
0.456
|
NM_001199319
NM_017929
|
PEX26
|
peroxisomal biogenesis factor 26
|
|
chr7_+_148823475
|
0.456
|
NM_020781
|
ZNF398
|
zinc finger protein 398
|
|
chr16_+_2802605
|
0.451
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr15_-_40987284
|
0.451
|
|
LOC100505648
|
uncharacterized LOC100505648
|
|
chr22_+_26825220
|
0.450
|
NM_020437
|
ASPHD2
|
aspartate beta-hydroxylase domain containing 2
|
|
chr16_-_15149827
|
0.447
|
NM_173474
|
NTAN1
|
N-terminal asparagine amidase
|
|
chr5_-_55008125
|
0.446
|
NM_173514
|
SLC38A9
|
solute carrier family 38, member 9
|
|
chr5_-_55008079
|
0.446
|
|
SLC38A9
|
solute carrier family 38, member 9
|
|
chr2_-_180871779
|
0.440
|
NM_020943
|
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chrX_-_106362018
|
0.440
|
|
RBM41
|
RNA binding motif protein 41
|
|
chrX_+_70797873
|
0.440
|
NM_052957
|
ACRC
|
acidic repeat containing
|
|
chr11_-_65314049
|
0.440
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr13_+_26828238
|
0.439
|
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr19_-_37263653
|
0.439
|
NM_001193552
|
ZNF850
|
zinc finger protein 850
|
|
chr16_-_30537852
|
0.438
|
NM_024671
|
ZNF768
|
zinc finger protein 768
|
|
chr12_+_88536045
|
0.438
|
NM_181783
|
TMTC3
|
transmembrane and tetratricopeptide repeat containing 3
|
|
chr1_+_93913566
|
0.437
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr9_-_138853007
|
0.433
|
|
UBAC1
|
UBA domain containing 1
|
|
chr16_+_2802656
|
0.432
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr3_-_186524483
|
0.429
|
NM_181573
|
RFC4
|
replication factor C (activator 1) 4, 37kDa
|
|
chr10_+_75541804
|
0.425
|
NM_203298
|
CHCHD1
|
coiled-coil-helix-coiled-coil-helix domain containing 1
|
|
chr4_+_148653452
|
0.422
|
NM_024605
|
ARHGAP10
|
Rho GTPase activating protein 10
|
|
chr15_+_85144219
|
0.420
|
NM_001007072
NM_017894
NM_181877
|
ZSCAN2
|
zinc finger and SCAN domain containing 2
|
|
chr11_-_82996898
|
0.416
|
|
CCDC90B
|
coiled-coil domain containing 90B
|
|
chrX_-_152939256
|
0.415
|
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr7_-_100888285
|
0.415
|
|
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae)
|
|
chr8_-_74884338
|
0.412
|
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C)
|
|
chr2_-_178483677
|
0.411
|
NM_152275
|
TTC30A
|
tetratricopeptide repeat domain 30A
|
|
chr6_-_84419018
|
0.411
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr9_+_130186652
|
0.409
|
NM_007135
|
ZNF79
|
zinc finger protein 79
|
|
chr12_-_102455739
|
0.409
|
|
CCDC53
|
coiled-coil domain containing 53
|
|
chr4_+_113218498
|
0.408
|
NM_001102406
NM_025144
|
ALPK1
|
alpha-kinase 1
|