|
chr6_-_32821592
|
7.373
|
|
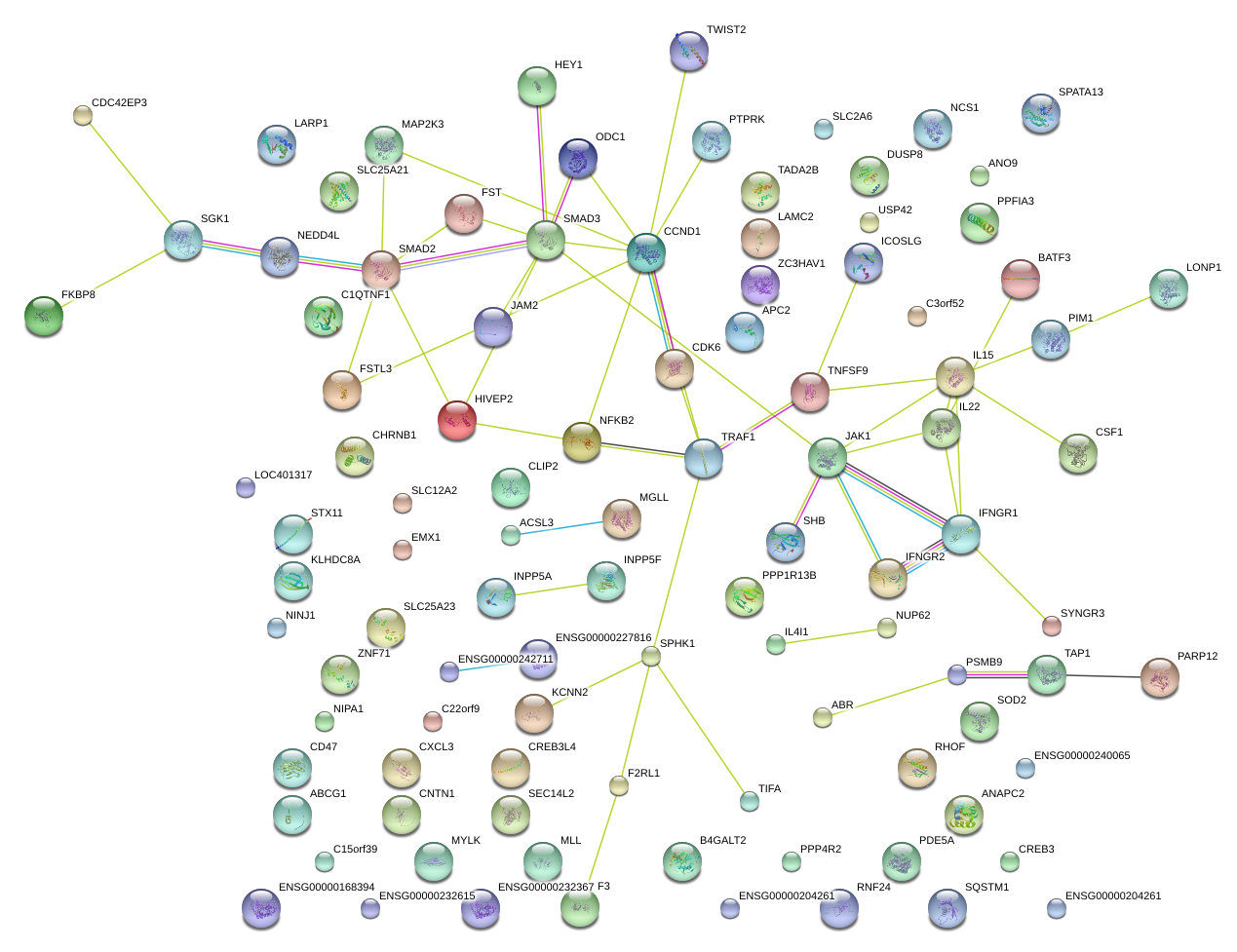
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr6_-_32821743
|
6.301
|
NM_000593
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr6_-_32821412
|
5.367
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr9_-_123691442
|
4.654
|
NM_001190945
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr6_+_32821924
|
4.623
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr3_-_127541160
|
3.872
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr21_+_27011766
|
3.532
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr6_+_37137882
|
3.466
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr21_+_27011588
|
3.394
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr4_+_142557722
|
3.306
|
NM_000585
NM_172175
|
IL15
|
interleukin 15
|
|
chr19_+_676346
|
3.176
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr9_+_35732207
|
3.004
|
NM_006368
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr21_+_43639202
|
2.925
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr9_+_35732629
|
2.751
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr7_+_73703704
|
2.710
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr10_+_104155431
|
2.644
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr16_+_2039945
|
2.574
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr3_+_111805174
|
2.300
|
NM_001171747
NM_024616
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr9_-_38069082
|
2.242
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr2_+_223725675
|
2.231
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr11_-_441995
|
2.220
|
NM_001012302
|
ANO9
|
anoctamin 9
|
|
chr3_-_127541975
|
2.144
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr1_-_65432203
|
2.137
|
|
JAK1
|
Janus kinase 1
|
|
chr17_-_1083008
|
2.093
|
NM_021962
|
ABR
|
active BCR-related gene
|
|
chr4_-_120549827
|
2.073
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr11_-_441956
|
2.059
|
|
ANO9
|
anoctamin 9
|
|
chr6_-_137113166
|
2.030
|
|
|
|
|
chr5_+_127419407
|
2.028
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr21_-_45660699
|
1.986
|
NM_015259
|
ICOSLG
|
inducible T-cell co-stimulator ligand
|
|
chr9_+_35732498
|
1.982
|
|
CREB3
|
cAMP responsive element binding protein 3
|
|
chr21_+_27012067
|
1.924
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr21_+_27012045
|
1.893
|
|
JAM2
|
junctional adhesion molecule 2
|
|
chr9_-_38069207
|
1.856
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr17_+_7348362
|
1.855
|
NM_000747
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle)
|
|
chr1_-_95007139
|
1.819
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr3_-_107809754
|
1.778
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr13_+_24734788
|
1.739
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr19_+_49622480
|
1.625
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr7_+_6144589
|
1.621
|
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr6_+_144471631
|
1.609
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr21_-_45660841
|
1.608
|
|
ICOSLG
|
inducible T-cell co-stimulator ligand
|
|
chr2_+_239756672
|
1.601
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr4_-_120550142
|
1.574
|
|
|
|
|
chr5_+_76114881
|
1.537
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr7_+_28448970
|
1.536
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr19_-_18654280
|
1.503
|
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr21_+_34775189
|
1.500
|
NM_005534
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1)
|
|
chr10_+_134351272
|
1.468
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr6_-_143266283
|
1.468
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr19_+_1446268
|
1.438
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr9_-_95896498
|
1.416
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr17_+_77020229
|
1.399
|
NM_030968
NM_198594
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr19_-_50432952
|
1.397
|
NM_016553
|
NUP62
|
nucleoporin 62kDa
|
|
chr9_-_136344182
|
1.388
|
NM_001145099
NM_017585
|
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chr20_-_3996035
|
1.388
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr17_+_21191347
|
1.387
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr1_-_212873306
|
1.363
|
NM_018664
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3
|
|
chr5_+_154135029
|
1.359
|
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr22_-_45608232
|
1.353
|
|
KIAA0930
|
KIAA0930
|
|
chr17_+_74380676
|
1.347
|
NM_001142601
NM_021972
NM_182965
|
SPHK1
|
sphingosine kinase 1
|
|
chr4_+_7045222
|
1.330
|
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr1_-_65431826
|
1.326
|
NM_002227
|
JAK1
|
Janus kinase 1
|
|
chr1_+_183155393
|
1.288
|
|
LAMC2
|
laminin, gamma 2
|
|
chr15_+_75494235
|
1.273
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr22_-_45608346
|
1.270
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr6_-_160114318
|
1.264
|
NM_000636
NM_001024465
NM_001024466
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr5_+_113769226
|
1.257
|
NM_170775
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr1_+_44445548
|
1.257
|
NM_030587
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr15_-_23086290
|
1.251
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr6_-_134496006
|
1.246
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr15_+_75494233
|
1.240
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr11_+_69455978
|
1.237
|
|
CCND1
|
cyclin D1
|
|
chr15_+_75494196
|
1.231
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr3_+_73046081
|
1.227
|
NM_174907
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr2_-_37899227
|
1.208
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr4_-_113206883
|
1.208
|
NM_052864
|
TIFA
|
TRAF-interacting protein with forkhead-associated domain
|
|
chr7_-_139762339
|
1.197
|
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr17_+_7348840
|
1.195
|
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle)
|
|
chr2_-_10588273
|
1.175
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr19_+_57106622
|
1.171
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr9_+_132934685
|
1.165
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr5_+_179234002
|
1.161
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr6_-_137540435
|
1.156
|
|
IFNGR1
|
interferon gamma receptor 1
|
|
chr18_+_55711746
|
1.150
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_+_110453456
|
1.147
|
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr2_-_37898768
|
1.146
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr11_+_118307072
|
1.141
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr4_-_74904329
|
1.140
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr6_-_128841432
|
1.128
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr18_+_55711890
|
1.109
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr14_-_104313730
|
1.109
|
NM_015316
|
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B
|
|
chr17_+_77020278
|
1.097
|
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr19_-_50432672
|
1.081
|
|
IL4I1
|
interleukin 4 induced 1
|
|
chr12_-_122231554
|
1.080
|
NM_019034
|
RHOF
|
ras homolog gene family, member F (in filopodia)
|
|
chr11_-_1593149
|
1.078
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr8_-_80679897
|
1.078
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr19_-_50432647
|
1.077
|
|
IL4I1
|
interleukin 4 induced 1
|
|
chr7_-_92465929
|
1.076
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr6_-_137540544
|
1.062
|
|
IFNGR1
|
interferon gamma receptor 1
|
|
chr7_-_138794393
|
1.047
|
NM_020119
NM_024625
|
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1
|
|
chr6_-_137540551
|
1.041
|
NM_000416
|
IFNGR1
|
interferon gamma receptor 1
|
|
chr7_-_92465876
|
1.040
|
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr6_-_137540504
|
1.039
|
|
IFNGR1
|
interferon gamma receptor 1
|
|
chr15_+_67430358
|
1.036
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr1_-_205313309
|
1.035
|
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr5_+_52776479
|
1.032
|
|
FST
|
follistatin
|
|
chr19_+_6531009
|
1.025
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr15_+_67358194
|
1.024
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr3_-_123603144
|
1.023
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr1_+_1072274
|
1.022
|
|
LOC254099
|
uncharacterized LOC254099
|
|
chr20_+_30946786
|
1.021
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr5_+_52776408
|
1.016
|
|
FST
|
follistatin
|
|
chr10_+_72575623
|
1.010
|
NM_003901
|
SGPL1
|
sphingosine-1-phosphate lyase 1
|
|
chr3_+_122399665
|
1.008
|
NM_017554
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr2_-_10588451
|
1.002
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr21_+_45736351
|
0.988
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr9_+_32384622
|
0.987
|
|
ACO1
|
aconitase 1, soluble
|
|
chr9_+_74764394
|
0.983
|
|
GDA
|
guanine deaminase
|
|
chr9_+_74764266
|
0.983
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr18_+_12308242
|
0.982
|
NM_032525
|
TUBB6
|
tubulin, beta 6 class V
|
|
chr8_+_104383785
|
0.981
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr3_-_182880463
|
0.970
|
NM_014398
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr19_-_17958773
|
0.970
|
|
JAK3
|
Janus kinase 3
|
|
chrX_-_119249774
|
0.970
|
NM_139282
|
RHOXF1
|
Rhox homeobox family, member 1
|
|
chr18_-_77793914
|
0.969
|
|
|
|
|
chr8_+_104383701
|
0.967
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr6_+_44095357
|
0.965
|
NM_018426
|
TMEM63B
|
transmembrane protein 63B
|
|
chr18_+_12308236
|
0.955
|
|
TUBB6
|
tubulin, beta 6 class V
|
|
chr7_-_2354010
|
0.954
|
|
SNX8
|
sorting nexin 8
|
|
chr6_-_160114186
|
0.952
|
|
SOD2
|
superoxide dismutase 2, mitochondrial
|
|
chr14_-_52535945
|
0.941
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr17_-_48207113
|
0.937
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr22_+_42470247
|
0.936
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr16_-_4466607
|
0.930
|
|
CORO7
|
coronin 7
|
|
chr7_+_21468293
|
0.922
|
|
|
|
|
chr1_+_44444848
|
0.919
|
NM_003780
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr9_+_74764382
|
0.917
|
|
GDA
|
guanine deaminase
|
|
chr3_-_158450205
|
0.917
|
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr16_+_88729730
|
0.916
|
|
MGC23284
|
uncharacterized LOC197187
|
|
chr12_-_49351251
|
0.914
|
NM_001659
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr3_+_10206562
|
0.912
|
NM_001570
|
IRAK2
|
interleukin-1 receptor-associated kinase 2
|
|
chr11_-_8739398
|
0.911
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr5_-_172756505
|
0.907
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr4_-_2758018
|
0.906
|
NM_024309
|
TNIP2
|
TNFAIP3 interacting protein 2
|
|
chr4_+_2420671
|
0.904
|
NM_001193282
|
LOC402160
|
uncharacterized LOC402160
|
|
chr6_+_108881010
|
0.897
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr14_+_51026853
|
0.892
|
|
ATL1
|
atlastin GTPase 1
|
|
chr3_+_53195214
|
0.892
|
NM_006254
NM_212539
|
PRKCD
|
protein kinase C, delta
|
|
chr22_+_41697452
|
0.891
|
NM_017590
|
ZC3H7B
|
zinc finger CCCH-type containing 7B
|
|
chr7_+_143058794
|
0.891
|
|
ZYX
|
zyxin
|
|
chr4_-_7436614
|
0.890
|
NM_001085382
|
PSAPL1
|
prosaposin-like 1 (gene/pseudogene)
|
|
chr2_+_150186498
|
0.881
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr6_-_46459710
|
0.880
|
NM_001251973
|
RCAN2
|
regulator of calcineurin 2
|
|
chr11_-_61735108
|
0.880
|
NM_002032
|
FTH1
|
ferritin, heavy polypeptide 1
|
|
chr11_+_67183144
|
0.880
|
NM_001166222
NM_020811
|
CARNS1
|
carnosine synthase 1
|
|
chr6_+_158402887
|
0.874
|
NM_003898
|
SYNJ2
|
synaptojanin 2
|
|
chr14_+_52118539
|
0.874
|
NM_152330
|
FRMD6
|
FERM domain containing 6
|
|
chr11_-_64645925
|
0.873
|
|
EHD1
|
EH-domain containing 1
|
|
chr4_+_74735107
|
0.859
|
NM_001511
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr6_+_135502477
|
0.859
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr11_-_128392061
|
0.856
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr21_+_45527253
|
0.854
|
|
PWP2
|
PWP2 periodic tryptophan protein homolog (yeast)
|
|
chr19_+_48828820
|
0.854
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr3_-_158450271
|
0.845
|
NM_002888
NM_206963
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1
|
|
chr5_-_9546157
|
0.842
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr20_+_45523247
|
0.842
|
NM_005244
NM_172110
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr21_+_45527202
|
0.825
|
|
PWP2
|
PWP2 periodic tryptophan protein homolog (yeast)
|
|
chrX_-_151143096
|
0.823
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr22_-_50893728
|
0.823
|
|
SBF1
|
SET binding factor 1
|
|
chr2_+_106361404
|
0.818
|
NM_001004722
NM_003581
|
NCK2
|
NCK adaptor protein 2
|
|
chr19_-_663155
|
0.815
|
|
RNF126
|
ring finger protein 126
|
|
chr9_-_136024522
|
0.813
|
NM_001042368
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr19_-_38720312
|
0.812
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr11_+_102188180
|
0.812
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr1_-_36851484
|
0.808
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr1_-_1051735
|
0.807
|
NM_017891
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr18_+_55711388
|
0.806
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_-_35888816
|
0.803
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr16_-_75498582
|
0.797
|
NM_145254
|
TMEM170A
|
transmembrane protein 170A
|
|
chr20_+_45523505
|
0.789
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr6_+_18155535
|
0.788
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr1_+_183155361
|
0.787
|
|
LAMC2
|
laminin, gamma 2
|
|
chr21_+_45719964
|
0.785
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr1_+_44444986
|
0.784
|
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr11_-_8932497
|
0.781
|
NM_005418
|
ST5
|
suppression of tumorigenicity 5
|
|
chr6_+_158402916
|
0.780
|
|
SYNJ2
|
synaptojanin 2
|
|
chr17_-_42907606
|
0.779
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr1_-_205290756
|
0.775
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr6_-_134495944
|
0.774
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_149865310
|
0.773
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr1_+_169075869
|
0.773
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr12_-_52779362
|
0.771
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr14_+_55034633
|
0.771
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr3_-_79068542
|
0.770
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr14_+_105155724
|
0.769
|
NM_001031714
NM_022489
NM_032714
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr1_-_27816667
|
0.767
|
|
WASF2
|
WAS protein family, member 2
|
|
chr6_-_44233244
|
0.766
|
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon
|
|
chr20_+_45523523
|
0.766
|
|
EYA2
|
eyes absent homolog 2 (Drosophila)
|
|
chr1_+_183155173
|
0.765
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr6_-_137113529
|
0.765
|
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|