Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
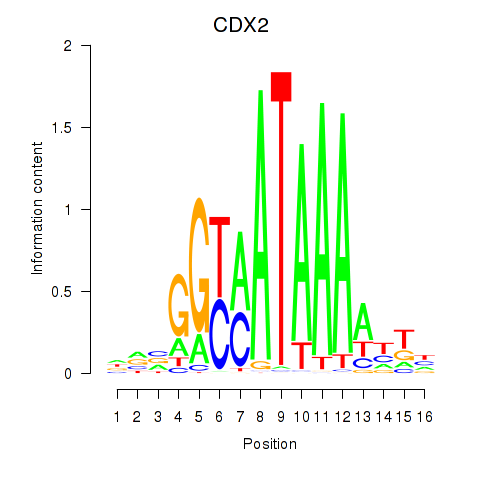
Results for CDX2
Z-value: 0.46
Transcription factors associated with CDX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX2
|
ENSG00000165556.9 | caudal type homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX2 | hg19_v2_chr13_-_28545276_28545276 | -0.35 | 8.2e-02 | Click! |
Activity profile of CDX2 motif
Sorted Z-values of CDX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_72649763 | 0.52 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr7_-_41742697 | 0.47 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr13_+_108922228 | 0.46 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr7_+_141811539 | 0.39 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr17_-_46690839 | 0.39 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr8_+_124084899 | 0.36 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr6_+_32812568 | 0.30 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_-_76957214 | 0.29 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr4_-_169239921 | 0.29 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr1_-_7913089 | 0.29 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr17_-_39646116 | 0.28 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr8_+_76452097 | 0.27 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr13_+_108921977 | 0.27 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr1_+_74701062 | 0.27 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr12_+_4130143 | 0.26 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr12_-_21927736 | 0.26 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr4_+_88754069 | 0.25 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_+_88754113 | 0.25 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr1_+_158979792 | 0.25 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_-_23886285 | 0.25 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr1_+_158979686 | 0.23 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr9_-_95640218 | 0.22 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr2_-_32390801 | 0.22 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr4_-_186317034 | 0.21 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr11_-_3663212 | 0.21 |
ENST00000397067.3
|
ART5
|
ADP-ribosyltransferase 5 |
| chr2_-_217236750 | 0.21 |
ENST00000273067.4
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
| chr8_+_16884740 | 0.20 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
| chr4_+_129349188 | 0.20 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr3_-_108672609 | 0.19 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr7_-_25268104 | 0.19 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr5_-_43515231 | 0.19 |
ENST00000306862.2
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr17_+_25936862 | 0.19 |
ENST00000582410.1
|
KSR1
|
kinase suppressor of ras 1 |
| chr3_-_108672742 | 0.19 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr7_-_130080977 | 0.18 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr5_+_121465207 | 0.18 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr1_+_109419596 | 0.18 |
ENST00000435987.1
ENST00000264126.3 |
GPSM2
|
G-protein signaling modulator 2 |
| chr9_-_86571628 | 0.18 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr19_-_48752812 | 0.18 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chrX_-_101771645 | 0.18 |
ENST00000289373.4
|
TMSB15A
|
thymosin beta 15a |
| chr6_+_158733692 | 0.17 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr22_-_32651326 | 0.17 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr19_-_48753104 | 0.17 |
ENST00000447740.2
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr8_+_145215928 | 0.17 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr6_+_25754927 | 0.17 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr15_+_35270552 | 0.16 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr19_+_21265028 | 0.16 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr7_-_16872932 | 0.15 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr7_-_26578407 | 0.15 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr16_-_80603558 | 0.15 |
ENST00000567317.1
|
RP11-18F14.1
|
RP11-18F14.1 |
| chr20_-_29994007 | 0.15 |
ENST00000376314.2
|
DEFB121
|
defensin, beta 121 |
| chr7_+_57509877 | 0.15 |
ENST00000420713.1
|
ZNF716
|
zinc finger protein 716 |
| chr11_+_120039685 | 0.15 |
ENST00000530303.1
ENST00000319763.1 |
AP000679.2
|
Uncharacterized protein |
| chr19_+_21264980 | 0.15 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr2_-_216878305 | 0.14 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr11_+_58390132 | 0.14 |
ENST00000361987.4
|
CNTF
|
ciliary neurotrophic factor |
| chr1_+_89246647 | 0.14 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr19_-_22018966 | 0.14 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr8_+_107738240 | 0.14 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr22_-_30642782 | 0.14 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr18_+_3252265 | 0.14 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr22_+_17956618 | 0.14 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr6_+_53948328 | 0.14 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr19_+_21106081 | 0.14 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr11_-_3400442 | 0.14 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr2_+_109204909 | 0.14 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_+_24645807 | 0.14 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr19_+_21579908 | 0.14 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr1_+_24645865 | 0.14 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_86848760 | 0.13 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr10_-_127505167 | 0.13 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr8_-_20040638 | 0.13 |
ENST00000519026.1
ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr4_-_76944621 | 0.13 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr4_+_146539415 | 0.13 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr2_-_233415220 | 0.13 |
ENST00000408957.3
|
TIGD1
|
tigger transposable element derived 1 |
| chr6_+_47666275 | 0.13 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr11_-_3400330 | 0.13 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
| chrX_+_64708615 | 0.13 |
ENST00000338957.4
ENST00000423889.3 |
ZC3H12B
|
zinc finger CCCH-type containing 12B |
| chr8_-_143859197 | 0.12 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr10_-_115614127 | 0.12 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr8_-_20040601 | 0.12 |
ENST00000265808.7
ENST00000522513.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr16_-_21663919 | 0.12 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr1_+_192127578 | 0.12 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr16_+_25228242 | 0.12 |
ENST00000219660.5
|
AQP8
|
aquaporin 8 |
| chr19_+_48497962 | 0.12 |
ENST00000596043.1
ENST00000597519.1 |
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr19_+_48497901 | 0.12 |
ENST00000339841.2
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr11_+_22688150 | 0.12 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr1_+_40942887 | 0.12 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr6_+_134274322 | 0.11 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr5_+_61874562 | 0.11 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr3_+_148545586 | 0.11 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr20_+_2276639 | 0.11 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr16_-_25122785 | 0.11 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr9_+_100174232 | 0.11 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr12_-_57146095 | 0.11 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr6_+_111408698 | 0.11 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr10_-_21661870 | 0.11 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr3_+_143690640 | 0.11 |
ENST00000315691.3
|
C3orf58
|
chromosome 3 open reading frame 58 |
| chr19_+_21579958 | 0.11 |
ENST00000339914.6
ENST00000599461.1 |
ZNF493
|
zinc finger protein 493 |
| chr18_+_3252206 | 0.10 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr17_+_58018269 | 0.10 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr22_+_32455111 | 0.10 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr5_-_148033726 | 0.10 |
ENST00000354217.2
ENST00000314512.6 ENST00000362016.2 |
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr1_+_241695424 | 0.10 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr12_-_18243075 | 0.09 |
ENST00000536890.1
|
RERGL
|
RERG/RAS-like |
| chr5_-_36001108 | 0.09 |
ENST00000333811.4
|
UGT3A1
|
UDP glycosyltransferase 3 family, polypeptide A1 |
| chr7_+_128784712 | 0.09 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr19_-_48759119 | 0.09 |
ENST00000522889.1
ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr5_+_135468516 | 0.09 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr11_+_71900572 | 0.09 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr1_+_220267429 | 0.09 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr9_+_100174344 | 0.09 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr2_-_40657397 | 0.09 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr15_+_40886199 | 0.08 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chr19_-_48753028 | 0.08 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr3_-_196911002 | 0.08 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr14_+_55494323 | 0.08 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr11_+_71900703 | 0.08 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr6_-_26189304 | 0.08 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr8_+_107738343 | 0.08 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr16_-_21431078 | 0.08 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr6_-_87804815 | 0.08 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chrX_+_65384182 | 0.08 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr3_+_63953415 | 0.08 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr15_-_41836441 | 0.08 |
ENST00000567866.1
ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1
|
RNA polymerase II associated protein 1 |
| chr15_+_67835517 | 0.07 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr14_+_52456327 | 0.07 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr9_+_131549610 | 0.07 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr12_-_52828147 | 0.07 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr1_-_106161540 | 0.07 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr7_+_64838786 | 0.07 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr3_-_38992052 | 0.07 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr4_+_174089904 | 0.07 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr14_+_52456193 | 0.07 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr5_-_58295712 | 0.07 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_+_128426535 | 0.07 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr3_-_18480260 | 0.07 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chrX_+_65384052 | 0.07 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr8_+_52730143 | 0.06 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr14_+_56584414 | 0.06 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr3_-_67705006 | 0.06 |
ENST00000492795.1
ENST00000493112.1 ENST00000307227.5 |
SUCLG2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr18_+_21032781 | 0.06 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr20_-_18810797 | 0.06 |
ENST00000278779.4
|
C20orf78
|
chromosome 20 open reading frame 78 |
| chr5_-_148442584 | 0.06 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr7_+_35756186 | 0.06 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr1_-_23495340 | 0.06 |
ENST00000418342.1
|
LUZP1
|
leucine zipper protein 1 |
| chr2_+_219081817 | 0.06 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr7_+_35756092 | 0.06 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr11_+_60048004 | 0.06 |
ENST00000532114.1
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr1_+_53480598 | 0.06 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr19_-_48753064 | 0.06 |
ENST00000520153.1
ENST00000357778.5 ENST00000520015.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr7_-_143105941 | 0.06 |
ENST00000275815.3
|
EPHA1
|
EPH receptor A1 |
| chr7_+_134464414 | 0.05 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr2_+_187371440 | 0.05 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr9_-_95055956 | 0.05 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr5_-_145562147 | 0.05 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr3_-_9921934 | 0.05 |
ENST00000423850.1
|
CIDEC
|
cell death-inducing DFFA-like effector c |
| chr3_+_107602030 | 0.05 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr4_+_186317133 | 0.05 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr5_-_148033693 | 0.05 |
ENST00000377888.3
ENST00000360693.3 |
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr7_+_64838712 | 0.05 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr6_-_127840336 | 0.05 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr5_+_177457525 | 0.05 |
ENST00000511856.1
ENST00000511189.1 |
FAM153C
|
family with sequence similarity 153, member C |
| chr19_+_4402659 | 0.05 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr10_-_129691195 | 0.05 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr10_+_85933494 | 0.05 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr3_-_156534754 | 0.05 |
ENST00000472943.1
ENST00000473352.1 |
LINC00886
|
long intergenic non-protein coding RNA 886 |
| chr1_+_46152886 | 0.04 |
ENST00000372025.4
|
TMEM69
|
transmembrane protein 69 |
| chr4_+_70894130 | 0.04 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr5_-_139937895 | 0.04 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr19_+_21106028 | 0.04 |
ENST00000597314.1
ENST00000601924.1 |
ZNF85
|
zinc finger protein 85 |
| chr5_+_175511859 | 0.04 |
ENST00000503724.2
ENST00000253490.4 |
FAM153B
|
family with sequence similarity 153, member B |
| chr5_+_72143988 | 0.04 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr5_+_98109322 | 0.04 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr1_+_210001309 | 0.04 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr9_-_95056010 | 0.04 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr12_+_64173583 | 0.04 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr2_-_151428735 | 0.04 |
ENST00000441356.1
|
AC104777.2
|
AC104777.2 |
| chr1_+_73771844 | 0.04 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr22_+_41956767 | 0.04 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr1_-_54355430 | 0.04 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr10_-_103599591 | 0.04 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr10_+_75504105 | 0.04 |
ENST00000535742.1
ENST00000546025.1 ENST00000345254.4 ENST00000540668.1 ENST00000339365.2 ENST00000411652.2 |
SEC24C
|
SEC24 family member C |
| chr6_+_125304502 | 0.03 |
ENST00000519799.1
ENST00000368414.2 ENST00000359704.2 |
RNF217
|
ring finger protein 217 |
| chr2_-_95787738 | 0.03 |
ENST00000272418.2
|
MRPS5
|
mitochondrial ribosomal protein S5 |
| chr2_+_27440229 | 0.03 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr14_+_53173890 | 0.03 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr17_+_7211656 | 0.03 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr8_-_13372395 | 0.03 |
ENST00000276297.4
ENST00000511869.1 |
DLC1
|
deleted in liver cancer 1 |
| chr12_-_118796910 | 0.03 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr14_+_53173910 | 0.03 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr2_+_138721850 | 0.03 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr16_+_14805546 | 0.03 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr16_+_68119764 | 0.03 |
ENST00000570212.1
ENST00000562926.1 |
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr17_-_72772462 | 0.03 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chrX_-_135962876 | 0.03 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr2_+_32390925 | 0.03 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr16_-_20338748 | 0.03 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr7_+_134464376 | 0.03 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr6_-_27782548 | 0.03 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr15_+_38746307 | 0.03 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.7 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.3 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.6 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0039534 | regulation of MDA-5 signaling pathway(GO:0039533) negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.6 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.6 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |