Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
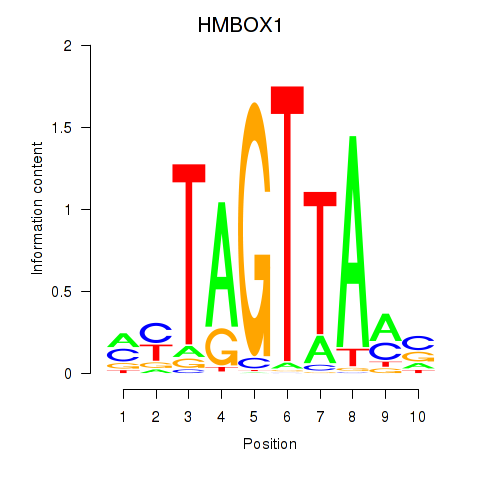
Results for HMBOX1
Z-value: 0.56
Transcription factors associated with HMBOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMBOX1
|
ENSG00000147421.13 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMBOX1 | hg19_v2_chr8_+_28747884_28747938 | -0.19 | 3.8e-01 | Click! |
Activity profile of HMBOX1 motif
Sorted Z-values of HMBOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_22234381 | 1.01 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_-_56236734 | 0.79 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr2_+_162087577 | 0.76 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr14_+_22615942 | 0.63 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr1_-_173176452 | 0.61 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr7_+_128095900 | 0.56 |
ENST00000435296.2
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr8_-_63951730 | 0.55 |
ENST00000260118.6
|
GGH
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr9_-_73029540 | 0.55 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr7_+_128095945 | 0.48 |
ENST00000257696.4
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr11_+_19798964 | 0.47 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr11_-_57194948 | 0.44 |
ENST00000533235.1
ENST00000526621.1 ENST00000352187.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr7_+_134576317 | 0.42 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr4_-_114900831 | 0.41 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr12_+_51633061 | 0.40 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr8_+_94929969 | 0.39 |
ENST00000517764.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr12_+_75874580 | 0.38 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr19_-_56826157 | 0.37 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr7_+_100728720 | 0.37 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr7_-_47579188 | 0.36 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr15_-_89764929 | 0.36 |
ENST00000268125.5
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr14_-_50999307 | 0.35 |
ENST00000013125.4
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr2_+_127413481 | 0.35 |
ENST00000259254.4
|
GYPC
|
glycophorin C (Gerbich blood group) |
| chr4_-_159080806 | 0.35 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr4_+_9172135 | 0.32 |
ENST00000512047.1
|
FAM90A26
|
family with sequence similarity 90, member A26 |
| chr19_+_16178317 | 0.31 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chr4_-_10117949 | 0.30 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr5_+_138629628 | 0.30 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chrX_+_70752945 | 0.30 |
ENST00000373701.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr20_+_43211149 | 0.30 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_+_206643787 | 0.29 |
ENST00000367120.3
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr8_+_66955648 | 0.29 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr1_+_109632425 | 0.28 |
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B |
| chr5_+_173316341 | 0.28 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr9_-_101984184 | 0.28 |
ENST00000476832.1
|
ALG2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr2_-_197226875 | 0.28 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr19_-_7553852 | 0.27 |
ENST00000593547.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr22_-_36635684 | 0.27 |
ENST00000358502.5
|
APOL2
|
apolipoprotein L, 2 |
| chr22_-_32334403 | 0.26 |
ENST00000543051.1
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr3_-_69101413 | 0.26 |
ENST00000398559.2
|
TMF1
|
TATA element modulatory factor 1 |
| chr1_+_89246647 | 0.25 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr11_+_73087309 | 0.25 |
ENST00000064780.2
ENST00000545687.1 |
RELT
|
RELT tumor necrosis factor receptor |
| chr5_-_133968459 | 0.24 |
ENST00000505758.1
ENST00000439578.1 ENST00000502286.1 |
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr16_-_30597000 | 0.24 |
ENST00000470110.1
ENST00000395216.2 |
ZNF785
|
zinc finger protein 785 |
| chr1_-_150669500 | 0.24 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_-_77272765 | 0.23 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr3_-_69101461 | 0.23 |
ENST00000543976.1
|
TMF1
|
TATA element modulatory factor 1 |
| chr19_-_7553889 | 0.23 |
ENST00000221480.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chrX_-_20236970 | 0.23 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr3_+_38347427 | 0.22 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr6_-_27114577 | 0.22 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr4_+_119810134 | 0.22 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr4_+_36283213 | 0.22 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr21_-_46340884 | 0.22 |
ENST00000302347.5
ENST00000517819.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chrX_+_70586140 | 0.22 |
ENST00000276072.3
|
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr5_+_140810132 | 0.22 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr19_-_8214730 | 0.21 |
ENST00000600128.1
|
FBN3
|
fibrillin 3 |
| chr7_+_134576151 | 0.21 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chrX_+_70503037 | 0.21 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr11_-_5207612 | 0.21 |
ENST00000380367.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr7_+_128379449 | 0.20 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr1_+_206643806 | 0.20 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr10_+_115312766 | 0.19 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr17_+_3118915 | 0.19 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr3_-_141747439 | 0.19 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chrX_+_70503433 | 0.18 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr14_+_31028348 | 0.18 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr12_+_117013656 | 0.18 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chrX_+_135730373 | 0.18 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr9_-_73477826 | 0.18 |
ENST00000396285.1
ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr14_+_50999744 | 0.18 |
ENST00000441560.2
|
ATL1
|
atlastin GTPase 1 |
| chrX_+_135730297 | 0.17 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr1_-_153283194 | 0.17 |
ENST00000290722.1
|
PGLYRP3
|
peptidoglycan recognition protein 3 |
| chr17_+_42925270 | 0.17 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chrM_+_5824 | 0.17 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr4_-_122302163 | 0.17 |
ENST00000394427.2
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chrX_-_30877837 | 0.17 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr15_+_69857515 | 0.17 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr16_-_18908196 | 0.17 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr20_+_68306 | 0.17 |
ENST00000382410.2
|
DEFB125
|
defensin, beta 125 |
| chr10_+_24755416 | 0.17 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr5_-_137667526 | 0.16 |
ENST00000503022.1
|
CDC25C
|
cell division cycle 25C |
| chrX_+_70752917 | 0.16 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr2_+_106468204 | 0.16 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr1_+_24286287 | 0.15 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr3_-_45838011 | 0.15 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr12_-_76879852 | 0.15 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr1_+_207943667 | 0.15 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr16_-_71842941 | 0.15 |
ENST00000423132.2
ENST00000433195.2 ENST00000569748.1 ENST00000570017.1 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr3_-_45837959 | 0.15 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr1_+_43803475 | 0.15 |
ENST00000372470.3
ENST00000413998.2 |
MPL
|
myeloproliferative leukemia virus oncogene |
| chr1_-_217250231 | 0.15 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chrX_-_15683147 | 0.15 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr6_+_29364416 | 0.15 |
ENST00000383555.2
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr1_+_175036966 | 0.15 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chrX_-_99987088 | 0.14 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr1_+_87595542 | 0.14 |
ENST00000461990.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr20_-_43133491 | 0.14 |
ENST00000411544.1
|
SERINC3
|
serine incorporator 3 |
| chr4_+_144303093 | 0.13 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_+_33155525 | 0.13 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein |
| chr6_-_107235287 | 0.13 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr8_+_30244580 | 0.13 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr2_+_68384976 | 0.13 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr12_+_72080253 | 0.12 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr2_+_159825143 | 0.12 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr5_+_140762268 | 0.12 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr7_-_28220354 | 0.12 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr17_-_55822653 | 0.12 |
ENST00000299415.2
|
AC007431.1
|
coiled-coil domain containing 182 |
| chr10_+_124913793 | 0.12 |
ENST00000368865.4
ENST00000538238.1 ENST00000368859.2 |
BUB3
|
BUB3 mitotic checkpoint protein |
| chr20_+_61584026 | 0.12 |
ENST00000370351.4
ENST00000370349.3 |
SLC17A9
|
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr1_+_27561104 | 0.12 |
ENST00000361771.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr19_-_18654293 | 0.12 |
ENST00000597547.1
ENST00000222308.4 ENST00000544835.3 ENST00000610101.1 ENST00000597960.3 ENST00000608443.1 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr1_-_167487758 | 0.12 |
ENST00000362089.5
|
CD247
|
CD247 molecule |
| chr11_-_58345569 | 0.12 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr6_+_134758827 | 0.12 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr1_-_150669604 | 0.12 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chrX_+_78426469 | 0.12 |
ENST00000276077.1
|
GPR174
|
G protein-coupled receptor 174 |
| chr1_+_87595497 | 0.11 |
ENST00000471417.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr19_-_35992780 | 0.11 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr1_-_217311090 | 0.11 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr13_-_108870623 | 0.11 |
ENST00000405925.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr21_+_45725050 | 0.11 |
ENST00000403390.1
|
PFKL
|
phosphofructokinase, liver |
| chr2_-_220110187 | 0.11 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr4_+_74301880 | 0.11 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr10_+_89419370 | 0.11 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr6_+_111303218 | 0.11 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr17_+_41177220 | 0.11 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr11_-_64825993 | 0.11 |
ENST00000340252.4
ENST00000355721.3 ENST00000356632.3 ENST00000355369.2 ENST00000339885.2 ENST00000358658.3 |
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr1_+_116654376 | 0.11 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr17_-_10452929 | 0.11 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr4_+_120860695 | 0.11 |
ENST00000505122.2
|
RP11-700N1.1
|
RP11-700N1.1 |
| chr2_-_214016314 | 0.11 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr2_+_182850551 | 0.11 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_-_220110111 | 0.11 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr2_+_234686976 | 0.10 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr6_-_107235331 | 0.10 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr1_+_222988363 | 0.10 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr1_+_27561007 | 0.10 |
ENST00000319394.3
|
WDTC1
|
WD and tetratricopeptide repeats 1 |
| chr17_+_26800648 | 0.10 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_-_6240183 | 0.10 |
ENST00000262450.3
ENST00000378021.1 |
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr3_+_38537960 | 0.10 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr2_+_192543153 | 0.10 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_+_26800756 | 0.10 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr12_-_62586543 | 0.10 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr2_+_36923933 | 0.10 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr12_-_54673871 | 0.10 |
ENST00000209875.4
|
CBX5
|
chromobox homolog 5 |
| chr7_-_102985288 | 0.10 |
ENST00000379263.3
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr20_+_42187682 | 0.09 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_-_57326704 | 0.09 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr8_+_107460147 | 0.09 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr10_-_112678904 | 0.09 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr6_+_167536230 | 0.09 |
ENST00000341935.5
ENST00000349984.4 |
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr5_+_138629417 | 0.09 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr20_+_42187608 | 0.09 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr6_+_26199737 | 0.09 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr16_-_71843047 | 0.09 |
ENST00000299980.4
ENST00000393512.3 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr1_-_152009460 | 0.09 |
ENST00000271638.2
|
S100A11
|
S100 calcium binding protein A11 |
| chr17_+_26800296 | 0.09 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_+_222988464 | 0.09 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr22_-_37880543 | 0.09 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr13_+_48611665 | 0.08 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr4_-_152149033 | 0.08 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr1_-_32384693 | 0.08 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr12_-_88423164 | 0.08 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr2_+_113825547 | 0.08 |
ENST00000341010.2
ENST00000337569.3 |
IL1F10
|
interleukin 1 family, member 10 (theta) |
| chr14_-_45431091 | 0.08 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr2_-_99952769 | 0.08 |
ENST00000409434.1
ENST00000434323.1 ENST00000264255.3 |
TXNDC9
|
thioredoxin domain containing 9 |
| chr14_+_75536280 | 0.08 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr1_+_196743943 | 0.08 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr12_+_49121213 | 0.08 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chrX_+_70586082 | 0.08 |
ENST00000373790.4
ENST00000449580.1 ENST00000423759.1 |
TAF1
|
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr17_-_29641084 | 0.08 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr17_+_3572087 | 0.07 |
ENST00000248378.5
ENST00000397133.2 |
EMC6
|
ER membrane protein complex subunit 6 |
| chr8_-_67874805 | 0.07 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr14_-_57272366 | 0.07 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr16_+_21689835 | 0.07 |
ENST00000286149.4
ENST00000388958.3 |
OTOA
|
otoancorin |
| chr14_+_75536335 | 0.07 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr10_+_112679301 | 0.07 |
ENST00000265277.5
ENST00000369452.4 |
SHOC2
|
soc-2 suppressor of clear homolog (C. elegans) |
| chr6_-_52668605 | 0.07 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr6_+_138725343 | 0.07 |
ENST00000607197.1
ENST00000367697.3 |
HEBP2
|
heme binding protein 2 |
| chr1_+_196743912 | 0.07 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr18_-_31803435 | 0.07 |
ENST00000589544.1
ENST00000269185.4 ENST00000261592.5 |
NOL4
|
nucleolar protein 4 |
| chr10_-_94257512 | 0.06 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr17_+_6544078 | 0.06 |
ENST00000250101.5
|
TXNDC17
|
thioredoxin domain containing 17 |
| chr12_-_10588539 | 0.06 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr2_+_127782787 | 0.06 |
ENST00000564121.1
|
RP11-521O16.2
|
RP11-521O16.2 |
| chrX_-_77225135 | 0.06 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr11_+_5474638 | 0.06 |
ENST00000341449.2
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2 |
| chr11_-_47447970 | 0.06 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chrX_+_37208540 | 0.06 |
ENST00000466533.1
ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1
TM4SF2
|
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr11_+_65769550 | 0.06 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chr19_+_55014085 | 0.06 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr17_-_3571934 | 0.06 |
ENST00000225525.3
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr6_+_30585486 | 0.06 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr12_-_53715328 | 0.06 |
ENST00000547757.1
ENST00000394384.3 ENST00000209873.4 |
AAAS
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr19_+_55014013 | 0.06 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr15_+_79166065 | 0.05 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr11_-_62572901 | 0.05 |
ENST00000439713.2
ENST00000531131.1 ENST00000530875.1 ENST00000531709.2 ENST00000294172.2 |
NXF1
|
nuclear RNA export factor 1 |
| chr6_+_27114861 | 0.05 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr1_+_109255279 | 0.05 |
ENST00000370017.3
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr20_-_48770244 | 0.05 |
ENST00000371650.5
ENST00000371652.4 ENST00000557021.1 |
TMEM189
|
transmembrane protein 189 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMBOX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 0.6 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 0.6 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 0.5 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.5 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.8 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.3 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.3 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 0.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.3 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 1.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |