Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
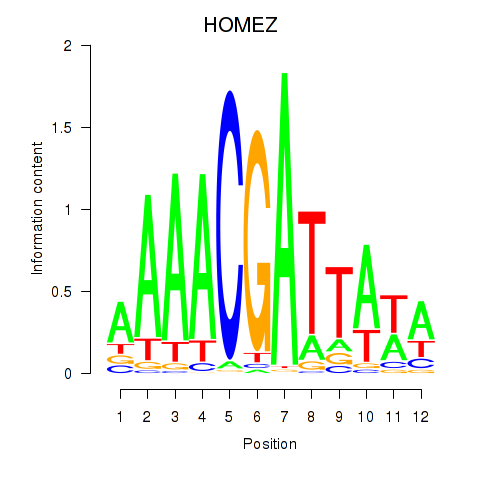
Results for HOMEZ
Z-value: 1.53
Transcription factors associated with HOMEZ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOMEZ
|
ENSG00000215271.6 | homeobox and leucine zipper encoding |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOMEZ | hg19_v2_chr14_-_23755297_23755350 | -0.48 | 1.6e-02 | Click! |
Activity profile of HOMEZ motif
Sorted Z-values of HOMEZ motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_156588249 | 7.85 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_+_156588350 | 7.80 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_+_156588115 | 7.71 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_+_156587979 | 7.01 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_-_70626314 | 6.89 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr16_-_30032610 | 6.77 |
ENST00000574405.1
|
DOC2A
|
double C2-like domains, alpha |
| chrX_-_119709637 | 5.51 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr7_+_89783689 | 4.88 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr4_+_126237554 | 4.07 |
ENST00000394329.3
|
FAT4
|
FAT atypical cadherin 4 |
| chr11_+_62104897 | 3.82 |
ENST00000415229.2
ENST00000535727.1 ENST00000301776.5 |
ASRGL1
|
asparaginase like 1 |
| chr9_+_71944241 | 3.64 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr4_-_186877806 | 3.32 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_47489240 | 3.26 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr12_+_10365404 | 3.21 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chrX_+_86772787 | 2.92 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr12_-_6055398 | 2.59 |
ENST00000327087.8
ENST00000356134.5 ENST00000546188.1 |
ANO2
|
anoctamin 2 |
| chr7_+_13141097 | 2.37 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr9_-_13279589 | 2.35 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr12_+_32655048 | 2.32 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_-_17229480 | 2.28 |
ENST00000532035.1
ENST00000540361.1 |
PIK3C2A
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr1_+_82266053 | 2.27 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr2_+_109237717 | 2.27 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_+_8995832 | 2.18 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr21_+_17443434 | 2.17 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr7_+_16793160 | 2.15 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chrX_+_86772707 | 2.12 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr1_+_109102652 | 2.09 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr19_-_18995029 | 2.02 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr9_-_13279406 | 2.00 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein |
| chr6_+_53659746 | 2.00 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr4_-_90759440 | 1.97 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr8_-_99129384 | 1.95 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr12_+_72233487 | 1.94 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr15_+_66585555 | 1.94 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr21_+_17443521 | 1.90 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_-_99129338 | 1.88 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr9_-_13279563 | 1.87 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr17_+_66521936 | 1.75 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr5_+_61874562 | 1.75 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr5_+_95066823 | 1.75 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr22_+_23046750 | 1.75 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr7_-_150020578 | 1.75 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chrX_+_102192200 | 1.74 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr1_-_68516393 | 1.70 |
ENST00000395201.1
|
DIRAS3
|
DIRAS family, GTP-binding RAS-like 3 |
| chr3_+_15643476 | 1.67 |
ENST00000436193.1
ENST00000383778.4 |
BTD
|
biotinidase |
| chr2_+_47630108 | 1.58 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr19_+_52901094 | 1.58 |
ENST00000391788.2
ENST00000436397.1 ENST00000391787.2 ENST00000360465.3 ENST00000494167.2 ENST00000493272.1 |
ZNF528
|
zinc finger protein 528 |
| chr2_+_191334212 | 1.58 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr6_+_35996859 | 1.58 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr7_+_101460882 | 1.57 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr5_+_73109339 | 1.56 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr2_-_190627481 | 1.56 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr2_+_120687335 | 1.56 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr13_+_39612485 | 1.54 |
ENST00000379599.2
|
NHLRC3
|
NHL repeat containing 3 |
| chr10_-_126849588 | 1.53 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr21_+_17909594 | 1.53 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr22_+_51112800 | 1.52 |
ENST00000414786.2
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr12_+_112451120 | 1.52 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr18_-_53069419 | 1.52 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr1_+_156561533 | 1.51 |
ENST00000368234.3
ENST00000368235.3 ENST00000368233.3 |
APOA1BP
|
apolipoprotein A-I binding protein |
| chr19_+_41698927 | 1.51 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr19_+_41284121 | 1.51 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr5_-_95158375 | 1.49 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr19_-_23869970 | 1.45 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr7_-_36406750 | 1.43 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr12_-_59314246 | 1.43 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr18_+_20513278 | 1.40 |
ENST00000327155.5
|
RBBP8
|
retinoblastoma binding protein 8 |
| chr13_+_39612442 | 1.40 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr12_-_10978957 | 1.38 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr13_+_32313658 | 1.38 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr5_+_1225470 | 1.34 |
ENST00000324642.3
ENST00000296821.4 |
SLC6A18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr14_-_105420241 | 1.34 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr2_-_99224915 | 1.33 |
ENST00000328709.3
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr4_+_108925787 | 1.32 |
ENST00000454409.2
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr1_+_196912902 | 1.30 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr1_+_97188188 | 1.29 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr22_-_29075853 | 1.28 |
ENST00000397906.2
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr17_-_36003487 | 1.24 |
ENST00000394367.3
ENST00000349699.2 |
DDX52
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
| chr5_+_140552218 | 1.23 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr2_-_112237835 | 1.23 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr4_-_163085107 | 1.22 |
ENST00000379164.4
|
FSTL5
|
follistatin-like 5 |
| chr12_+_60058458 | 1.22 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr19_+_35417939 | 1.22 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr12_-_65153175 | 1.20 |
ENST00000543646.1
ENST00000542058.1 ENST00000258145.3 |
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr2_+_87769459 | 1.19 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr4_-_99064387 | 1.15 |
ENST00000295268.3
|
STPG2
|
sperm-tail PG-rich repeat containing 2 |
| chr21_+_17553910 | 1.15 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_112450915 | 1.14 |
ENST00000437003.2
ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116
|
transmembrane protein 116 |
| chrX_-_80457385 | 1.14 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr8_-_93978309 | 1.13 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr1_+_212208919 | 1.11 |
ENST00000366991.4
ENST00000542077.1 |
DTL
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr4_-_110723194 | 1.11 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr5_-_156569850 | 1.11 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr20_-_35402123 | 1.11 |
ENST00000373740.3
ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1
|
DSN1, MIS12 kinetochore complex component |
| chr4_-_163085141 | 1.10 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr1_-_62190793 | 1.09 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chrX_-_134429952 | 1.08 |
ENST00000370764.1
|
ZNF75D
|
zinc finger protein 75D |
| chr1_+_20208870 | 1.05 |
ENST00000375120.3
|
OTUD3
|
OTU domain containing 3 |
| chr12_-_8803128 | 1.04 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr16_-_4588822 | 1.03 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr8_+_86089460 | 1.03 |
ENST00000418930.2
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chrX_+_54834004 | 1.03 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr8_-_66474884 | 1.02 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr17_+_37856253 | 1.01 |
ENST00000540147.1
ENST00000584450.1 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr5_+_64920826 | 1.01 |
ENST00000438419.2
ENST00000231526.4 ENST00000505553.1 ENST00000545191.1 |
TRAPPC13
|
trafficking protein particle complex 13 |
| chr17_+_37856299 | 1.00 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr3_-_194373831 | 1.00 |
ENST00000437613.1
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr6_-_109702885 | 0.97 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr11_+_114168085 | 0.97 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr15_-_26108355 | 0.96 |
ENST00000356865.6
|
ATP10A
|
ATPase, class V, type 10A |
| chr5_+_57878859 | 0.96 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr12_+_124155652 | 0.96 |
ENST00000426174.2
ENST00000303372.5 |
TCTN2
|
tectonic family member 2 |
| chr19_+_21541777 | 0.94 |
ENST00000380870.4
ENST00000597810.1 ENST00000594245.1 |
ZNF738
|
zinc finger protein 738 |
| chr19_+_41119323 | 0.92 |
ENST00000599724.1
ENST00000597071.1 ENST00000243562.9 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr6_+_44214824 | 0.91 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr1_-_182360918 | 0.91 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr9_-_86536323 | 0.91 |
ENST00000297814.2
ENST00000413982.1 ENST00000334204.2 |
KIF27
|
kinesin family member 27 |
| chr2_+_103089756 | 0.91 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr6_-_116575226 | 0.90 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr12_+_12878829 | 0.90 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr1_+_163039143 | 0.90 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr2_-_178129853 | 0.89 |
ENST00000397062.3
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chrX_-_64196351 | 0.89 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr16_-_5115913 | 0.88 |
ENST00000474471.3
|
C16orf89
|
chromosome 16 open reading frame 89 |
| chr7_+_150020363 | 0.88 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr3_-_137851220 | 0.88 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr2_+_66666432 | 0.86 |
ENST00000495021.2
|
MEIS1
|
Meis homeobox 1 |
| chrX_-_64196307 | 0.85 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr14_-_45722605 | 0.85 |
ENST00000310806.4
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr4_+_54243917 | 0.85 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr6_+_80816342 | 0.84 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr2_-_86850949 | 0.84 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr3_-_156878540 | 0.83 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr8_+_28748765 | 0.83 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr15_+_65843130 | 0.83 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr8_-_93978346 | 0.82 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr5_-_175388327 | 0.82 |
ENST00000432305.2
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr8_-_93978333 | 0.81 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr9_+_104296122 | 0.81 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr11_+_120207787 | 0.81 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr17_-_39183452 | 0.80 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr11_-_130184555 | 0.80 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr19_-_52408257 | 0.80 |
ENST00000354957.3
ENST00000600738.1 ENST00000595418.1 ENST00000599530.1 |
ZNF649
|
zinc finger protein 649 |
| chr5_-_64920115 | 0.80 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr8_-_93978357 | 0.78 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr12_+_100594557 | 0.78 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr10_-_126849068 | 0.78 |
ENST00000494626.2
ENST00000337195.5 |
CTBP2
|
C-terminal binding protein 2 |
| chr1_+_163038565 | 0.77 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr9_-_21228221 | 0.77 |
ENST00000413767.2
|
IFNA17
|
interferon, alpha 17 |
| chr19_+_21324863 | 0.76 |
ENST00000598331.1
|
ZNF431
|
zinc finger protein 431 |
| chr3_-_114343039 | 0.75 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr9_+_99690592 | 0.74 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr8_-_124749609 | 0.74 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr22_+_40573921 | 0.74 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr8_+_86099884 | 0.73 |
ENST00000517476.1
ENST00000521429.1 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr12_-_71551868 | 0.73 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr3_-_138312971 | 0.73 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr4_+_48807155 | 0.72 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr2_-_45838374 | 0.72 |
ENST00000263736.4
|
SRBD1
|
S1 RNA binding domain 1 |
| chr5_-_52405564 | 0.72 |
ENST00000510818.2
ENST00000396954.3 ENST00000508922.1 ENST00000361377.4 ENST00000582677.1 ENST00000584946.1 ENST00000450852.3 |
MOCS2
|
molybdenum cofactor synthesis 2 |
| chr1_-_54405773 | 0.71 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_-_15114603 | 0.71 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr7_+_99905325 | 0.71 |
ENST00000332397.6
ENST00000437326.2 |
SPDYE3
|
speedy/RINGO cell cycle regulator family member E3 |
| chr21_+_17442799 | 0.70 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr15_+_77223960 | 0.70 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr21_+_34638656 | 0.69 |
ENST00000290200.2
|
IL10RB
|
interleukin 10 receptor, beta |
| chr1_+_32608566 | 0.68 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr19_+_16059818 | 0.68 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor, family 10, subfamily H, member 4 |
| chr2_-_201729284 | 0.68 |
ENST00000434813.2
|
CLK1
|
CDC-like kinase 1 |
| chr2_-_8977714 | 0.67 |
ENST00000319688.5
ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220
|
kinase D-interacting substrate, 220kDa |
| chr11_-_7985193 | 0.67 |
ENST00000328600.2
|
NLRP10
|
NLR family, pyrin domain containing 10 |
| chr9_-_95166884 | 0.67 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr12_-_71551652 | 0.67 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr17_+_36584662 | 0.67 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr19_-_47616992 | 0.66 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr12_-_55375622 | 0.66 |
ENST00000316577.8
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr7_-_56119156 | 0.66 |
ENST00000421312.1
ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr4_+_47487285 | 0.66 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr11_+_101983176 | 0.65 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr14_-_106967788 | 0.64 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr1_+_215740709 | 0.64 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr2_+_219246746 | 0.64 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr1_+_203595903 | 0.63 |
ENST00000367218.3
ENST00000367219.3 ENST00000391954.2 |
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr2_-_3523507 | 0.62 |
ENST00000327435.6
|
ADI1
|
acireductone dioxygenase 1 |
| chr1_+_174843548 | 0.62 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_-_49466686 | 0.62 |
ENST00000273598.3
ENST00000436744.2 |
NICN1
|
nicolin 1 |
| chr7_-_48019101 | 0.62 |
ENST00000432627.1
ENST00000258774.5 ENST00000432325.1 ENST00000446009.1 |
HUS1
|
HUS1 checkpoint homolog (S. pombe) |
| chrX_-_64196376 | 0.62 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr13_-_31038370 | 0.61 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr14_+_50234309 | 0.61 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr7_-_99332719 | 0.61 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr19_-_55677920 | 0.61 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr14_+_58765103 | 0.60 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr11_+_18194384 | 0.60 |
ENST00000314254.3
|
MRGPRX4
|
MAS-related GPR, member X4 |
| chr11_+_43380459 | 0.60 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr16_-_5116025 | 0.60 |
ENST00000472572.3
ENST00000315997.5 ENST00000422873.1 ENST00000350219.4 |
C16orf89
|
chromosome 16 open reading frame 89 |
| chr2_+_234545148 | 0.60 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr1_+_153950202 | 0.60 |
ENST00000608236.1
|
RP11-422P24.11
|
RP11-422P24.11 |
| chr4_+_83956237 | 0.60 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr13_-_31736478 | 0.60 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOMEZ
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 30.4 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 1.0 | 4.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.8 | 2.4 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.5 | 1.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.5 | 4.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 3.8 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.4 | 2.0 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.4 | 1.5 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.4 | 1.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.4 | 1.1 | GO:0002859 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.3 | 1.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 1.6 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.3 | 1.6 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.3 | 5.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.3 | 2.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 | 1.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 8.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.3 | 2.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.7 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.2 | 0.9 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 0.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.7 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 0.6 | GO:0070839 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.2 | 1.7 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.6 | GO:0033242 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.2 | 0.6 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.2 | 0.8 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 0.7 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.2 | 0.4 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.4 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 0.9 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 1.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 3.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.7 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 2.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 0.8 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.2 | 1.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.6 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 | 0.6 | GO:0002424 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.4 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.1 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 2.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 1.1 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.1 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 0.4 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.1 | 0.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 1.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 6.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.3 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.1 | 0.3 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.6 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.4 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 3.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.4 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.1 | 0.3 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 1.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 1.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.9 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 1.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 1.7 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 2.3 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.1 | 1.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.8 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 1.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.6 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 1.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.4 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 0.5 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.8 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.7 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 1.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:0015811 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 1.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 1.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.7 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.1 | 0.7 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.7 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 1.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.6 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 2.3 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 0.9 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 2.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 2.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.3 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 1.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 1.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.4 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.7 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 1.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.4 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 1.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.5 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 1.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 1.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.6 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.4 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.9 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.9 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.9 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 2.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.6 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.1 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 1.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.3 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 1.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 29.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.8 | 6.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 1.6 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.4 | 1.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.3 | 6.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 0.7 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 0.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.2 | 1.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 2.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.7 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.0 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 3.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 2.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 6.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 4.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.0 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 4.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.5 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 4.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.6 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 2.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 6.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 1.1 | 29.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.6 | 4.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 1.6 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.4 | 1.7 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.4 | 2.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 2.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 0.9 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.3 | 1.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 2.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 0.8 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.3 | 6.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 1.0 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.2 | 0.7 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 0.9 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 2.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 0.6 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 0.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.2 | 0.6 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.2 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 0.8 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 1.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 3.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 0.9 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 2.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 1.6 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 3.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 1.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.7 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 1.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 2.6 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.6 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 1.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 7.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.4 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.8 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 1.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.4 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.2 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 4.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.6 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 1.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 5.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 7.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 4.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.0 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 29.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 6.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 3.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |