Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HOXA10_HOXB9
Z-value: 0.59
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
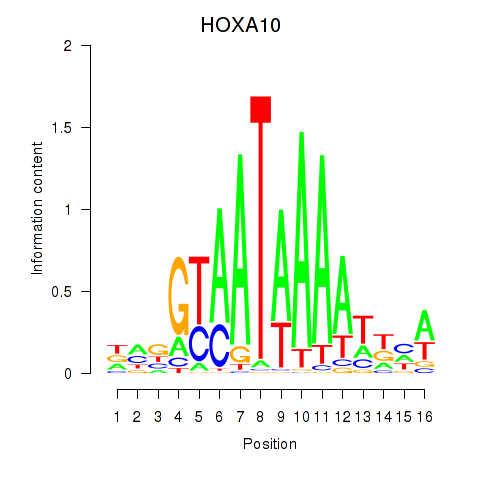
HOXA10
|
ENSG00000253293.3 | homeobox A10 |
|
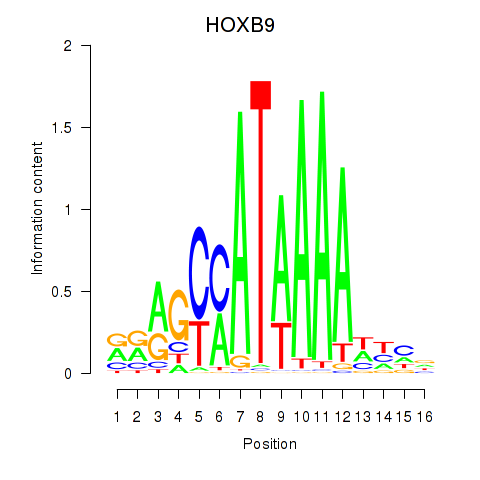
HOXB9
|
ENSG00000170689.8 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg19_v2_chr7_-_27213893_27213954 | -0.38 | 6.2e-02 | Click! |
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | 0.11 | 5.9e-01 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_67418047 | 1.85 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr15_-_55541227 | 1.53 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr14_-_72458326 | 1.22 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr15_-_58571445 | 0.73 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr6_+_28249299 | 0.72 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr6_+_28249332 | 0.71 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr21_-_35899113 | 0.63 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr2_+_161993465 | 0.63 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr3_-_150690471 | 0.60 |
ENST00000468836.1
ENST00000328863.4 |
CLRN1
|
clarin 1 |
| chr18_-_5396271 | 0.60 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr1_-_160231451 | 0.58 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr5_-_16738451 | 0.55 |
ENST00000274203.9
ENST00000515803.1 |
MYO10
|
myosin X |
| chr2_+_208423840 | 0.53 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr2_-_216878305 | 0.52 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr4_+_70916119 | 0.48 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr22_-_32651326 | 0.46 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr2_+_173686303 | 0.45 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr5_+_135496675 | 0.45 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr2_+_208423891 | 0.45 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr16_-_75498553 | 0.42 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chrX_-_117119243 | 0.40 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr6_-_137539651 | 0.40 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr15_+_63414760 | 0.39 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr9_+_82187630 | 0.37 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_+_82187487 | 0.37 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_+_82188077 | 0.36 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr14_+_31046959 | 0.36 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr10_+_102106829 | 0.36 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr2_+_161993412 | 0.34 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr3_-_172241250 | 0.32 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr10_+_78078088 | 0.32 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr18_-_52989217 | 0.31 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr10_-_129691195 | 0.31 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr8_-_81083890 | 0.31 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr9_+_34652164 | 0.30 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr7_+_108210012 | 0.30 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr16_-_15180257 | 0.29 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr3_+_138340067 | 0.28 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_+_1800179 | 0.28 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr1_+_151735431 | 0.27 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr14_-_51027838 | 0.27 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_-_6420930 | 0.26 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr1_-_161207875 | 0.26 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr18_-_52989525 | 0.26 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr12_-_71551868 | 0.26 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr18_-_12656715 | 0.25 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr1_-_161207953 | 0.25 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr2_+_102928009 | 0.25 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr1_-_161208013 | 0.24 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr15_-_98417780 | 0.24 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr1_-_161207986 | 0.23 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr2_-_209010874 | 0.23 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr14_-_60952739 | 0.23 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr11_-_13517565 | 0.23 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr21_+_35736302 | 0.22 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr3_-_50375657 | 0.22 |
ENST00000395126.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr12_-_71551652 | 0.21 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr3_+_138340049 | 0.21 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_+_54892550 | 0.20 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr14_+_56584414 | 0.20 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr1_+_192127578 | 0.20 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr18_-_53069419 | 0.19 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr18_-_74839891 | 0.19 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr7_-_140482926 | 0.19 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr3_+_57875711 | 0.18 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr13_-_103719196 | 0.18 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr6_+_12717892 | 0.18 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr10_-_24770632 | 0.18 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr12_-_81992111 | 0.17 |
ENST00000443686.3
ENST00000407050.4 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chrX_+_55744228 | 0.17 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr2_+_220144052 | 0.17 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chrX_-_45060135 | 0.17 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chrX_+_55744166 | 0.17 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr16_-_80603558 | 0.17 |
ENST00000567317.1
|
RP11-18F14.1
|
RP11-18F14.1 |
| chr6_+_88054530 | 0.16 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr17_-_67264947 | 0.16 |
ENST00000586811.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr2_+_220143989 | 0.16 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr18_+_42260861 | 0.16 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr7_+_35756186 | 0.16 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr7_+_35756092 | 0.16 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chrX_+_105937068 | 0.16 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr4_-_69536346 | 0.15 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr4_-_72649763 | 0.15 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr15_+_66797627 | 0.15 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chrX_+_144908928 | 0.15 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr11_-_7961141 | 0.15 |
ENST00000360759.3
|
OR10A3
|
olfactory receptor, family 10, subfamily A, member 3 |
| chr1_+_84630645 | 0.15 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_+_91933726 | 0.15 |
ENST00000534113.2
|
SECISBP2
|
SECIS binding protein 2 |
| chr4_+_56815102 | 0.15 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr5_+_67586465 | 0.14 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_-_38821373 | 0.14 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr9_+_105757590 | 0.14 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr19_-_3500635 | 0.14 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr19_+_21106028 | 0.14 |
ENST00000597314.1
ENST00000601924.1 |
ZNF85
|
zinc finger protein 85 |
| chr4_-_90759440 | 0.14 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_-_111312622 | 0.14 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr20_-_25320367 | 0.14 |
ENST00000450393.1
ENST00000491682.1 |
ABHD12
|
abhydrolase domain containing 12 |
| chr4_-_110723194 | 0.14 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr2_-_231989808 | 0.13 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chrX_+_36254051 | 0.13 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr1_-_226926864 | 0.13 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr6_-_83903600 | 0.13 |
ENST00000506587.1
ENST00000507554.1 |
PGM3
|
phosphoglucomutase 3 |
| chr7_+_56131917 | 0.12 |
ENST00000434526.2
ENST00000275607.9 ENST00000395435.2 ENST00000413952.2 ENST00000342190.6 ENST00000437307.2 ENST00000413756.1 ENST00000451338.1 |
SUMF2
|
sulfatase modifying factor 2 |
| chr15_+_93447675 | 0.12 |
ENST00000536619.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr3_-_112329110 | 0.12 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr3_+_157828152 | 0.12 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr14_+_73706308 | 0.12 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr6_-_52859046 | 0.12 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr3_-_196911002 | 0.12 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr16_+_70380825 | 0.12 |
ENST00000417604.2
|
DDX19A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
| chr15_-_56209306 | 0.12 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr6_+_32812568 | 0.12 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr20_-_8000426 | 0.12 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chrX_+_36246735 | 0.11 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr6_+_80816342 | 0.11 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr3_+_149191723 | 0.11 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chrX_-_80457385 | 0.11 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr1_+_110993795 | 0.11 |
ENST00000271331.3
|
PROK1
|
prokineticin 1 |
| chr15_+_52155001 | 0.11 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr17_+_61086917 | 0.11 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr20_-_7921090 | 0.11 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr3_-_100712352 | 0.11 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr6_+_83903061 | 0.11 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr11_-_114430570 | 0.11 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr20_-_50384864 | 0.10 |
ENST00000311637.5
ENST00000402822.1 |
ATP9A
|
ATPase, class II, type 9A |
| chr16_-_21663919 | 0.10 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr5_+_141346385 | 0.10 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr2_+_109237717 | 0.10 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_+_90198535 | 0.10 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr5_-_87516448 | 0.10 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr1_+_151739131 | 0.10 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr9_+_21967137 | 0.10 |
ENST00000441769.2
|
C9orf53
|
chromosome 9 open reading frame 53 |
| chr2_+_109204743 | 0.10 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr7_+_138915102 | 0.09 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr19_+_50016610 | 0.09 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr22_-_23922448 | 0.09 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr17_-_39324424 | 0.09 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr4_-_104021009 | 0.09 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr14_+_57671888 | 0.09 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr5_+_72143988 | 0.09 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr11_-_3400330 | 0.09 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
| chr4_+_186317133 | 0.09 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr10_-_103599591 | 0.09 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr6_-_27782548 | 0.08 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr15_+_66797455 | 0.08 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr2_-_71062938 | 0.08 |
ENST00000410009.3
|
CD207
|
CD207 molecule, langerin |
| chr11_+_120973375 | 0.08 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr1_+_248201474 | 0.08 |
ENST00000366479.2
|
OR2L2
|
olfactory receptor, family 2, subfamily L, member 2 |
| chr12_-_18243075 | 0.08 |
ENST00000536890.1
|
RERGL
|
RERG/RAS-like |
| chr1_-_17766198 | 0.08 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr4_+_88896819 | 0.08 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr5_-_86534822 | 0.08 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr3_-_9921934 | 0.08 |
ENST00000423850.1
|
CIDEC
|
cell death-inducing DFFA-like effector c |
| chr1_-_186430222 | 0.08 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr8_-_59412717 | 0.08 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr11_-_3400442 | 0.08 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr10_+_24755416 | 0.08 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr17_-_39150385 | 0.08 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr12_-_8765446 | 0.08 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr4_+_155484103 | 0.07 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr1_+_229440129 | 0.07 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr21_+_34398153 | 0.07 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chrX_-_100604184 | 0.07 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr20_-_29994007 | 0.07 |
ENST00000376314.2
|
DEFB121
|
defensin, beta 121 |
| chr2_+_101591314 | 0.07 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr6_+_44215603 | 0.07 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr15_+_40886439 | 0.07 |
ENST00000532056.1
ENST00000399668.2 |
CASC5
|
cancer susceptibility candidate 5 |
| chr5_-_176433693 | 0.07 |
ENST00000507513.1
ENST00000511320.1 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chrX_-_15619076 | 0.06 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr3_+_15045419 | 0.06 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr6_-_87804815 | 0.06 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr4_+_70894130 | 0.06 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr9_+_124413873 | 0.06 |
ENST00000408936.3
|
DAB2IP
|
DAB2 interacting protein |
| chr16_-_21663950 | 0.06 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr12_-_12491608 | 0.06 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr5_+_140767452 | 0.06 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr4_-_186317034 | 0.06 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr6_-_109804412 | 0.06 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr13_+_24144509 | 0.06 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr1_-_155880672 | 0.06 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr5_-_125930929 | 0.06 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr10_-_73848531 | 0.05 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr11_-_104916034 | 0.05 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr8_-_81083731 | 0.05 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr20_+_42984330 | 0.05 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr1_+_52682052 | 0.05 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr16_+_70380732 | 0.05 |
ENST00000302243.7
|
DDX19A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
| chr22_-_43010928 | 0.05 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr10_-_73848086 | 0.05 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr18_+_19192228 | 0.05 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr14_+_22433675 | 0.05 |
ENST00000390442.3
|
TRAV12-3
|
T cell receptor alpha variable 12-3 |
| chrX_+_70503433 | 0.05 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr2_-_51259229 | 0.05 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr6_-_31830655 | 0.05 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr8_+_107738343 | 0.05 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr2_-_51259292 | 0.05 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr9_+_130026756 | 0.05 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr11_-_40315640 | 0.05 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr19_+_21265028 | 0.05 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr1_+_154401791 | 0.05 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 1.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 1.0 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.2 | 1.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.2 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.4 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.1 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.0 | 0.4 | GO:0071786 | nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 1.1 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.0 | 0.3 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0035726 | regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.4 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.5 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.3 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 1.0 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 1.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 0.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |