Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
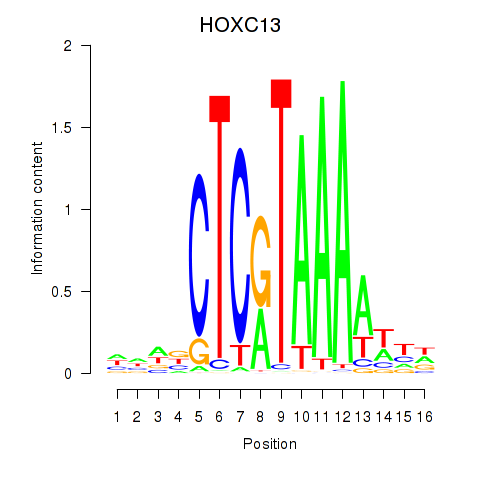
Results for HOXC13
Z-value: 0.54
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.3 | homeobox C13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg19_v2_chr12_+_54332535_54332636 | -0.14 | 5.2e-01 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_112127981 | 1.85 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr2_+_161993465 | 0.73 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr9_-_74675521 | 0.69 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr11_+_118175132 | 0.67 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr2_+_162016827 | 0.65 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_150208320 | 0.60 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr21_-_27423339 | 0.58 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr11_+_30252549 | 0.56 |
ENST00000254122.3
ENST00000417547.1 |
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr2_-_177502254 | 0.54 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr21_-_46012386 | 0.53 |
ENST00000400368.1
|
KRTAP10-6
|
keratin associated protein 10-6 |
| chr12_+_45609862 | 0.44 |
ENST00000423947.3
ENST00000435642.1 |
ANO6
|
anoctamin 6 |
| chr2_+_161993412 | 0.43 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr12_+_45609797 | 0.43 |
ENST00000425752.2
|
ANO6
|
anoctamin 6 |
| chr6_-_11807277 | 0.42 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr2_+_162016804 | 0.39 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr12_-_47473425 | 0.38 |
ENST00000550413.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr16_-_68033356 | 0.36 |
ENST00000393847.1
ENST00000573808.1 ENST00000572624.1 |
DPEP2
|
dipeptidase 2 |
| chr2_+_162016916 | 0.33 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_3496171 | 0.33 |
ENST00000399756.4
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr16_-_46865286 | 0.31 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr3_-_155461515 | 0.29 |
ENST00000399242.2
|
AC104472.1
|
CDNA FLJ26134 fis, clone TMS03713; Uncharacterized protein |
| chr17_+_56769924 | 0.27 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr13_+_34392185 | 0.26 |
ENST00000380071.3
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr14_+_96722539 | 0.26 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr1_+_109656719 | 0.25 |
ENST00000457623.2
ENST00000529753.1 |
KIAA1324
|
KIAA1324 |
| chr12_+_96883347 | 0.25 |
ENST00000524981.4
ENST00000298953.3 |
C12orf55
|
chromosome 12 open reading frame 55 |
| chr2_+_241499719 | 0.25 |
ENST00000405954.1
|
DUSP28
|
dual specificity phosphatase 28 |
| chr1_-_150208291 | 0.24 |
ENST00000533654.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr15_-_58571445 | 0.24 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_+_134551583 | 0.24 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr5_+_134181625 | 0.22 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chr1_-_166845515 | 0.22 |
ENST00000367874.4
|
TADA1
|
transcriptional adaptor 1 |
| chr10_+_94352956 | 0.22 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr1_+_109656532 | 0.21 |
ENST00000531664.1
ENST00000534476.1 |
KIAA1324
|
KIAA1324 |
| chrX_-_30993201 | 0.21 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr1_+_174128639 | 0.21 |
ENST00000251507.4
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_54243917 | 0.20 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr4_-_128887069 | 0.20 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr1_-_150208363 | 0.20 |
ENST00000436748.2
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_+_141178440 | 0.20 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr1_+_84630645 | 0.19 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_109656579 | 0.19 |
ENST00000526264.1
ENST00000369939.3 |
KIAA1324
|
KIAA1324 |
| chr1_+_179050512 | 0.19 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr6_-_130031358 | 0.19 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr17_+_66521936 | 0.18 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr6_+_151042224 | 0.18 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_231560790 | 0.18 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr19_-_8809139 | 0.18 |
ENST00000324436.3
|
ACTL9
|
actin-like 9 |
| chr11_-_85565906 | 0.18 |
ENST00000544076.1
|
AP000974.1
|
CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein |
| chr13_-_20077417 | 0.18 |
ENST00000382978.1
ENST00000400230.2 ENST00000255310.6 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr10_+_13628933 | 0.18 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr3_+_111630451 | 0.18 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr7_+_80275621 | 0.17 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr14_-_50319482 | 0.17 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr1_+_174844645 | 0.17 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_-_64023410 | 0.17 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr17_+_48450575 | 0.17 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr4_+_122722466 | 0.17 |
ENST00000243498.5
ENST00000379663.3 ENST00000509800.1 |
EXOSC9
|
exosome component 9 |
| chr18_+_3466248 | 0.17 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr7_+_23749945 | 0.16 |
ENST00000354639.3
ENST00000531170.1 ENST00000444333.2 ENST00000428484.1 |
STK31
|
serine/threonine kinase 31 |
| chr4_+_170581213 | 0.16 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr2_+_201390843 | 0.16 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr8_-_27850141 | 0.15 |
ENST00000524352.1
|
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr7_+_23749767 | 0.15 |
ENST00000355870.3
|
STK31
|
serine/threonine kinase 31 |
| chr4_-_148605265 | 0.15 |
ENST00000541232.1
ENST00000322396.6 |
PRMT10
|
protein arginine methyltransferase 10 (putative) |
| chr7_+_106505696 | 0.15 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_+_78511586 | 0.15 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr19_+_38085731 | 0.15 |
ENST00000589117.1
|
ZNF540
|
zinc finger protein 540 |
| chr1_-_150208412 | 0.15 |
ENST00000532744.1
ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr19_+_21265028 | 0.15 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr20_-_18810797 | 0.15 |
ENST00000278779.4
|
C20orf78
|
chromosome 20 open reading frame 78 |
| chr2_+_138721850 | 0.14 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr1_+_219347203 | 0.14 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr14_-_50319758 | 0.14 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr3_+_101498269 | 0.14 |
ENST00000491511.2
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr7_+_80275953 | 0.13 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_-_150208498 | 0.13 |
ENST00000314136.8
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_-_58571935 | 0.13 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_+_102514019 | 0.13 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chr4_-_150736962 | 0.13 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr19_+_3762645 | 0.13 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr7_+_106505912 | 0.13 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr7_-_32534850 | 0.13 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_+_148508845 | 0.13 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr2_+_61372226 | 0.13 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr7_-_27169801 | 0.13 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr16_-_11367452 | 0.13 |
ENST00000327157.2
|
PRM3
|
protamine 3 |
| chr3_+_122513642 | 0.12 |
ENST00000261038.5
|
DIRC2
|
disrupted in renal carcinoma 2 |
| chr7_-_16872932 | 0.12 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr4_+_118955500 | 0.12 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr15_+_79166065 | 0.12 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr11_+_61891445 | 0.12 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr1_-_246580705 | 0.12 |
ENST00000541742.1
|
SMYD3
|
SET and MYND domain containing 3 |
| chr3_-_123339418 | 0.12 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr4_+_128702969 | 0.12 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr21_+_47706537 | 0.12 |
ENST00000397691.1
|
YBEY
|
ybeY metallopeptidase (putative) |
| chr14_+_24701819 | 0.12 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_+_219347186 | 0.12 |
ENST00000366928.5
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr22_+_38142235 | 0.11 |
ENST00000407319.2
ENST00000403663.2 ENST00000428075.1 |
TRIOBP
|
TRIO and F-actin binding protein |
| chr9_+_40028620 | 0.11 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr1_+_158979680 | 0.11 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr14_+_24701628 | 0.11 |
ENST00000355299.4
ENST00000559836.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_+_158979686 | 0.11 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr19_+_45409011 | 0.11 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr4_+_88720698 | 0.11 |
ENST00000226284.5
|
IBSP
|
integrin-binding sialoprotein |
| chr4_-_170533723 | 0.11 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr14_+_73634537 | 0.11 |
ENST00000406768.1
|
PSEN1
|
presenilin 1 |
| chr18_-_5544241 | 0.11 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_-_123339343 | 0.11 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr12_-_11287243 | 0.11 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr16_-_18908196 | 0.11 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr19_-_38085633 | 0.11 |
ENST00000593133.1
ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571
|
zinc finger protein 571 |
| chr16_-_2379688 | 0.11 |
ENST00000567910.1
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr2_+_48010312 | 0.10 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr1_-_85097431 | 0.10 |
ENST00000327308.3
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr3_+_148545586 | 0.10 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr2_+_102953608 | 0.10 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr1_+_50571949 | 0.10 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_-_17555164 | 0.10 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr4_+_70861647 | 0.10 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chrX_+_13707235 | 0.10 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr15_+_92006567 | 0.10 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr1_-_220220000 | 0.10 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr14_+_72052983 | 0.10 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr14_+_24702099 | 0.10 |
ENST00000420554.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr17_-_39538550 | 0.10 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr14_+_24702073 | 0.10 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr2_+_172778952 | 0.10 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr5_-_146781153 | 0.10 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr15_+_69854027 | 0.10 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr17_-_56769382 | 0.09 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr4_-_164395014 | 0.09 |
ENST00000280605.3
|
TKTL2
|
transketolase-like 2 |
| chr3_-_179169181 | 0.09 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr12_-_91539918 | 0.09 |
ENST00000548218.1
|
DCN
|
decorin |
| chr16_+_12059050 | 0.09 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr4_-_17513702 | 0.09 |
ENST00000428702.2
ENST00000508623.1 ENST00000513615.1 |
QDPR
|
quinoid dihydropteridine reductase |
| chr2_-_225811747 | 0.09 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr21_+_25801041 | 0.09 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr5_-_79287060 | 0.09 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr8_+_118147498 | 0.08 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr10_+_70847852 | 0.08 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr7_+_80275752 | 0.08 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_+_206858232 | 0.08 |
ENST00000294981.4
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr1_+_90460661 | 0.08 |
ENST00000340281.4
ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326
|
zinc finger protein 326 |
| chr1_-_109656439 | 0.08 |
ENST00000369949.4
|
C1orf194
|
chromosome 1 open reading frame 194 |
| chr19_+_21264980 | 0.08 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr6_+_29141311 | 0.08 |
ENST00000377167.2
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2 |
| chr10_+_90354503 | 0.08 |
ENST00000531458.1
|
LIPJ
|
lipase, family member J |
| chr7_+_23749894 | 0.07 |
ENST00000433467.2
|
STK31
|
serine/threonine kinase 31 |
| chr6_-_51952418 | 0.07 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr5_+_169010638 | 0.07 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr11_+_62037622 | 0.07 |
ENST00000227918.2
ENST00000525380.1 |
SCGB2A2
|
secretoglobin, family 2A, member 2 |
| chr6_-_146057144 | 0.07 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr10_-_79398250 | 0.07 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr19_+_7011509 | 0.07 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr4_-_17513851 | 0.07 |
ENST00000281243.5
|
QDPR
|
quinoid dihydropteridine reductase |
| chr1_+_46769303 | 0.07 |
ENST00000311672.5
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr5_+_138678131 | 0.07 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr9_-_5304432 | 0.07 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr16_+_86612112 | 0.07 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr14_+_24702127 | 0.07 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_-_112106556 | 0.07 |
ENST00000443498.1
|
ADORA3
|
adenosine A3 receptor |
| chr4_-_155533787 | 0.07 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr1_+_55107449 | 0.07 |
ENST00000421030.2
ENST00000545244.1 ENST00000339553.5 ENST00000409996.1 ENST00000454855.2 |
MROH7
|
maestro heat-like repeat family member 7 |
| chr10_-_69455873 | 0.07 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr11_-_114430570 | 0.07 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr2_+_33359687 | 0.07 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_89413456 | 0.07 |
ENST00000500381.2
|
RP11-13A1.1
|
RP11-13A1.1 |
| chr7_+_90012986 | 0.07 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr11_+_22689648 | 0.07 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr1_-_168464875 | 0.07 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr1_+_246887349 | 0.06 |
ENST00000366510.3
|
SCCPDH
|
saccharopine dehydrogenase (putative) |
| chr22_+_23101182 | 0.06 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr2_+_138722028 | 0.06 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chrM_+_8527 | 0.06 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr2_-_37384175 | 0.06 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr14_-_106725723 | 0.06 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr5_-_64920115 | 0.06 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr10_-_51623203 | 0.06 |
ENST00000444743.1
ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr4_-_175204765 | 0.06 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr7_-_30066233 | 0.06 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr1_+_153631130 | 0.06 |
ENST00000368685.5
|
SNAPIN
|
SNAP-associated protein |
| chr6_+_88182643 | 0.06 |
ENST00000369556.3
ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr22_-_19435209 | 0.06 |
ENST00000546308.1
ENST00000541063.1 ENST00000399568.1 ENST00000333059.5 |
HIRA
C22orf39
|
histone cell cycle regulator chromosome 22 open reading frame 39 |
| chr13_-_103451307 | 0.06 |
ENST00000376004.4
|
KDELC1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr17_-_17480779 | 0.06 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr17_-_39140549 | 0.06 |
ENST00000377755.4
|
KRT40
|
keratin 40 |
| chr9_-_69229650 | 0.06 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr2_+_48796120 | 0.06 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr11_+_94706804 | 0.05 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr2_+_33359646 | 0.05 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_-_165424973 | 0.05 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr1_+_248402231 | 0.05 |
ENST00000306687.1
|
OR2M4
|
olfactory receptor, family 2, subfamily M, member 4 |
| chrX_+_49687216 | 0.05 |
ENST00000376088.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr10_+_51371390 | 0.05 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr6_-_33239712 | 0.05 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr11_-_102576537 | 0.05 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr11_+_7618413 | 0.05 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr10_-_79398127 | 0.05 |
ENST00000372443.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_86174065 | 0.05 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 0.9 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.7 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.6 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.3 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.2 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 0.4 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.3 | GO:0032252 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:0051344 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 1.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.5 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.0 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.5 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.6 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 1.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0030914 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.5 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.6 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 1.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |