Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for HOXD9
Z-value: 0.88
Transcription factors associated with HOXD9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD9
|
ENSG00000128709.10 | homeobox D9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD9 | hg19_v2_chr2_+_176987088_176987088 | -0.44 | 2.8e-02 | Click! |
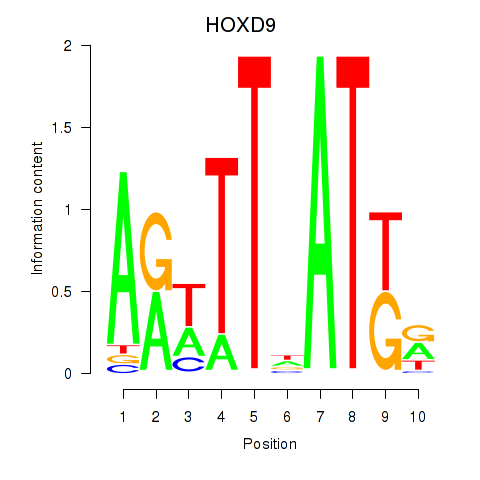
Activity profile of HOXD9 motif
Sorted Z-values of HOXD9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76944621 | 7.91 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr1_-_173020056 | 4.64 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr6_+_32812568 | 4.13 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_+_101185290 | 3.60 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr4_+_74606223 | 3.58 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr18_+_61554932 | 3.34 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr2_-_231989808 | 2.69 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr2_+_143635067 | 2.66 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chrX_+_144908928 | 2.49 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr18_-_5396271 | 2.20 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr2_-_202562716 | 2.06 |
ENST00000428900.2
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr2_-_202562774 | 1.99 |
ENST00000396886.3
ENST00000409143.1 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr22_-_41682172 | 1.93 |
ENST00000356244.3
|
RANGAP1
|
Ran GTPase activating protein 1 |
| chr7_-_92777606 | 1.91 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr11_-_128457446 | 1.73 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr3_-_107777208 | 1.57 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr3_-_27498235 | 1.40 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr12_-_88974236 | 1.35 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr3_+_111393501 | 1.32 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_-_104905840 | 1.30 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr7_-_107880508 | 1.22 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr6_+_132891461 | 1.20 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chrX_-_68385274 | 1.19 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr18_-_57027194 | 1.13 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr2_+_161993465 | 1.12 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_104972158 | 1.12 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr16_-_18887627 | 1.08 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chrX_-_68385354 | 1.08 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr6_+_149539767 | 1.03 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr7_-_121944491 | 0.99 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr15_+_66797627 | 0.99 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr4_-_76928641 | 0.96 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr22_+_23487513 | 0.95 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr7_-_122339162 | 0.94 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr19_+_10397648 | 0.86 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr15_-_55562582 | 0.86 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_95391315 | 0.84 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr4_-_68749699 | 0.84 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chrX_-_110513703 | 0.81 |
ENST00000324068.1
|
CAPN6
|
calpain 6 |
| chr2_-_175712270 | 0.81 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr11_+_61957687 | 0.80 |
ENST00000306238.3
|
SCGB1D1
|
secretoglobin, family 1D, member 1 |
| chr2_+_161993412 | 0.79 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr4_+_175839551 | 0.78 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr21_+_35736302 | 0.76 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr6_+_123317116 | 0.75 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr22_+_30477000 | 0.73 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr6_-_88411911 | 0.73 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr8_+_86089460 | 0.72 |
ENST00000418930.2
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr8_+_86089619 | 0.68 |
ENST00000256117.5
ENST00000416274.2 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr3_-_157221380 | 0.68 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_143635222 | 0.68 |
ENST00000375773.2
ENST00000409512.1 ENST00000410015.2 |
KYNU
|
kynureninase |
| chr15_+_66797455 | 0.66 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr17_-_10452929 | 0.65 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr2_+_114647504 | 0.64 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr6_+_29274403 | 0.64 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr4_-_68749745 | 0.64 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr4_+_186990298 | 0.61 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr1_+_181057638 | 0.61 |
ENST00000367577.4
|
IER5
|
immediate early response 5 |
| chr3_-_167191814 | 0.59 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr11_-_104827425 | 0.58 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_30253410 | 0.57 |
ENST00000533718.1
|
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr6_+_45390222 | 0.56 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr3_-_182703688 | 0.56 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr2_+_185463093 | 0.56 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr6_+_15401075 | 0.55 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr1_-_236046872 | 0.53 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr6_+_13272904 | 0.50 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr21_-_19775973 | 0.50 |
ENST00000284885.3
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr2_+_143886877 | 0.50 |
ENST00000295095.6
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr3_+_155838337 | 0.50 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_+_149887090 | 0.49 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr9_+_12693336 | 0.49 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr19_+_55014085 | 0.47 |
ENST00000351841.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr15_+_52155001 | 0.46 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr8_-_20040601 | 0.46 |
ENST00000265808.7
ENST00000522513.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr9_+_96846740 | 0.45 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_130423200 | 0.45 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr8_-_20040638 | 0.45 |
ENST00000519026.1
ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr19_+_55014013 | 0.45 |
ENST00000301202.2
|
LAIR2
|
leukocyte-associated immunoglobulin-like receptor 2 |
| chr12_-_21927736 | 0.44 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr3_-_195310802 | 0.44 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr5_+_135496675 | 0.44 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr2_-_228497888 | 0.44 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr4_+_71263599 | 0.44 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr6_+_131894284 | 0.44 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr19_-_56826157 | 0.43 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr5_+_147582387 | 0.43 |
ENST00000325630.2
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr20_+_5987890 | 0.43 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_-_100643765 | 0.42 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chrX_-_15332665 | 0.41 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr5_+_94727048 | 0.41 |
ENST00000283357.5
|
FAM81B
|
family with sequence similarity 81, member B |
| chr3_+_108855558 | 0.40 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr14_+_22615942 | 0.40 |
ENST00000390457.2
|
TRAV27
|
T cell receptor alpha variable 27 |
| chr12_+_26348246 | 0.39 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr10_-_31146615 | 0.38 |
ENST00000444692.2
|
ZNF438
|
zinc finger protein 438 |
| chr10_-_115614127 | 0.38 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr22_-_36236265 | 0.38 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_-_75748115 | 0.37 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_-_163695238 | 0.37 |
ENST00000328032.4
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr6_-_31632962 | 0.37 |
ENST00000456540.1
ENST00000445768.1 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr5_-_177207634 | 0.37 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr1_+_146373546 | 0.36 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr12_+_15125954 | 0.36 |
ENST00000266395.2
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr1_-_165414414 | 0.35 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr1_-_244006528 | 0.35 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr13_+_46039037 | 0.34 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr2_+_89986318 | 0.34 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr3_+_63428982 | 0.34 |
ENST00000479198.1
ENST00000460711.1 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr17_-_73389737 | 0.34 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr4_-_146859787 | 0.34 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr2_-_163695128 | 0.34 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr7_+_5920429 | 0.34 |
ENST00000242104.5
|
OCM
|
oncomodulin |
| chr1_+_192127578 | 0.33 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr6_+_31021225 | 0.32 |
ENST00000565192.1
ENST00000562344.1 |
HCG22
|
HLA complex group 22 |
| chr6_+_131958436 | 0.32 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr1_-_146082633 | 0.31 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr6_+_10528560 | 0.31 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chrX_-_130423386 | 0.31 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr2_+_145780739 | 0.31 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr20_-_13971255 | 0.31 |
ENST00000284951.5
ENST00000378072.5 |
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr11_+_49050504 | 0.30 |
ENST00000332682.7
|
TRIM49B
|
tripartite motif containing 49B |
| chr2_-_179567206 | 0.30 |
ENST00000414766.1
|
TTN
|
titin |
| chrX_-_109590174 | 0.30 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chrX_+_135388147 | 0.28 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr8_+_7801144 | 0.28 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr8_+_104892639 | 0.28 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_104615595 | 0.27 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr9_-_123812542 | 0.27 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr11_-_13517565 | 0.27 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chrX_+_48620147 | 0.26 |
ENST00000303227.6
|
GLOD5
|
glyoxalase domain containing 5 |
| chr1_+_87012753 | 0.26 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr11_+_12115543 | 0.26 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr11_-_16430399 | 0.26 |
ENST00000528252.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_128284020 | 0.26 |
ENST00000295321.4
ENST00000455721.2 |
IWS1
|
IWS1 homolog (S. cerevisiae) |
| chr14_+_62462541 | 0.26 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr15_-_54025300 | 0.26 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr7_-_130353553 | 0.26 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr2_+_145780725 | 0.26 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr3_-_112564797 | 0.25 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr2_+_145780767 | 0.25 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr5_+_125967372 | 0.24 |
ENST00000357147.3
|
C5orf48
|
chromosome 5 open reading frame 48 |
| chr4_-_168155577 | 0.24 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr19_+_7030589 | 0.24 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3-like 5 |
| chr1_+_160370344 | 0.23 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr15_-_90222610 | 0.23 |
ENST00000300055.5
|
PLIN1
|
perilipin 1 |
| chr22_+_50609150 | 0.23 |
ENST00000159647.5
ENST00000395842.2 |
PANX2
|
pannexin 2 |
| chr3_+_94657086 | 0.23 |
ENST00000463200.1
|
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr22_-_32651326 | 0.22 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr15_-_90222642 | 0.22 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chr4_-_83765613 | 0.22 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr8_+_107738343 | 0.22 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr4_-_168155730 | 0.22 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr5_+_177435986 | 0.21 |
ENST00000398106.2
|
FAM153C
|
family with sequence similarity 153, member C |
| chr10_+_115614370 | 0.21 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr10_+_5406935 | 0.21 |
ENST00000380433.3
|
UCN3
|
urocortin 3 |
| chr1_-_39339777 | 0.21 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr7_-_92855762 | 0.21 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr17_+_66521936 | 0.21 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr3_+_63428752 | 0.20 |
ENST00000295894.5
|
SYNPR
|
synaptoporin |
| chr5_+_29143603 | 0.20 |
ENST00000602470.1
ENST00000602801.1 |
CTD-2118P12.1
|
CTD-2118P12.1 |
| chr6_+_53948328 | 0.20 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr6_+_1312675 | 0.20 |
ENST00000296839.2
|
FOXQ1
|
forkhead box Q1 |
| chr1_+_197237352 | 0.20 |
ENST00000538660.1
ENST00000367400.3 ENST00000367399.2 |
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr12_-_95510743 | 0.19 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr14_+_57671888 | 0.19 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr6_+_63921351 | 0.19 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr13_-_47012325 | 0.19 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr3_+_94657118 | 0.19 |
ENST00000466089.1
ENST00000470465.1 |
LINC00879
|
long intergenic non-protein coding RNA 879 |
| chr1_+_45140360 | 0.19 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr12_+_9980069 | 0.19 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr7_+_93535866 | 0.19 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr1_+_44870866 | 0.19 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr2_-_136594740 | 0.18 |
ENST00000264162.2
|
LCT
|
lactase |
| chr2_-_158345462 | 0.18 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chrY_-_13524717 | 0.18 |
ENST00000331172.6
|
SLC9B1P1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 pseudogene 1 |
| chr11_-_105010320 | 0.18 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr5_+_147582348 | 0.18 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr1_-_152332480 | 0.17 |
ENST00000388718.5
|
FLG2
|
filaggrin family member 2 |
| chr1_+_240177627 | 0.17 |
ENST00000447095.1
|
FMN2
|
formin 2 |
| chr4_+_48485341 | 0.17 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr4_-_168155700 | 0.16 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_-_39646116 | 0.16 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr1_+_196857144 | 0.16 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr8_-_66701319 | 0.16 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr14_+_23654525 | 0.15 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr8_-_26724784 | 0.15 |
ENST00000380573.3
|
ADRA1A
|
adrenoceptor alpha 1A |
| chr22_-_37213554 | 0.15 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr18_-_70532906 | 0.15 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr14_-_68000442 | 0.15 |
ENST00000554278.1
|
TMEM229B
|
transmembrane protein 229B |
| chr9_-_138391692 | 0.15 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr1_+_202385953 | 0.14 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_-_10607084 | 0.14 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr17_+_60447579 | 0.14 |
ENST00000450662.2
|
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr10_-_28623368 | 0.14 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr8_-_117886955 | 0.14 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr3_+_155860751 | 0.14 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr15_+_58430567 | 0.14 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr3_+_102153859 | 0.14 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr6_-_49712123 | 0.14 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr7_+_93535817 | 0.14 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 1.1 | 3.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.9 | 8.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.6 | 2.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 2.7 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.5 | 3.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 1.3 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 1.9 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 2.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.7 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 1.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 | 0.8 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 1.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 1.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 0.6 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.4 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.9 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.8 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 1.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.4 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 1.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 3.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.7 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.3 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.4 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.2 | GO:0061341 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.2 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 1.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.4 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.5 | GO:0048023 | regulation of melanin biosynthetic process(GO:0048021) positive regulation of melanin biosynthetic process(GO:0048023) regulation of secondary metabolite biosynthetic process(GO:1900376) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 1.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 4.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.3 | GO:2000197 | regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.0 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 1.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 1.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of activated T cell proliferation(GO:0046007) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.5 | 4.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 2.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.3 | 1.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.9 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 2.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 8.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.6 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.6 | 3.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.5 | 3.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.9 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 0.9 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 2.7 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 1.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 3.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.4 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.4 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.4 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 4.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 4.6 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 8.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 3.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.7 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 12.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 3.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.6 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 4.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 4.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 3.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |