Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
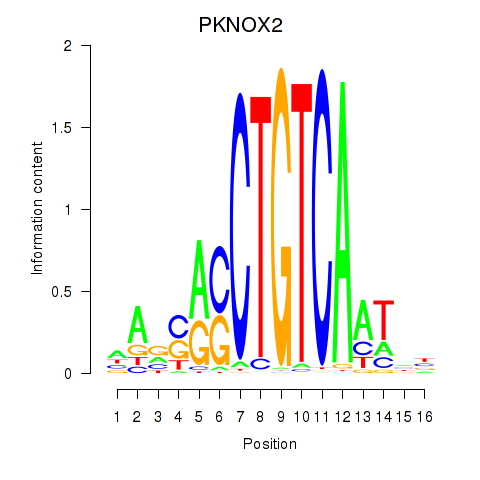
Results for PKNOX2
Z-value: 0.48
Transcription factors associated with PKNOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX2
|
ENSG00000165495.11 | PBX/knotted 1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX2 | hg19_v2_chr11_+_125034586_125034604 | -0.14 | 5.0e-01 | Click! |
Activity profile of PKNOX2 motif
Sorted Z-values of PKNOX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_157892167 | 1.25 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr4_-_157892055 | 1.01 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr4_-_157892498 | 0.90 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr4_+_128703295 | 0.60 |
ENST00000296464.4
ENST00000508549.1 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr6_+_138188551 | 0.56 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr4_-_149365827 | 0.55 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr16_+_2880296 | 0.55 |
ENST00000571723.1
|
ZG16B
|
zymogen granule protein 16B |
| chr16_+_2880254 | 0.55 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chrX_-_39923656 | 0.49 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr6_-_37225391 | 0.45 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr6_-_101329157 | 0.43 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr8_+_31496809 | 0.38 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr12_+_69201923 | 0.35 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr5_-_58882219 | 0.33 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr6_-_101329191 | 0.31 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr1_+_74701062 | 0.31 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr6_-_11382478 | 0.28 |
ENST00000397378.3
ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr16_+_2880369 | 0.28 |
ENST00000572863.1
|
ZG16B
|
zymogen granule protein 16B |
| chr11_-_111741994 | 0.27 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr16_+_2880157 | 0.26 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr1_+_181057638 | 0.26 |
ENST00000367577.4
|
IER5
|
immediate early response 5 |
| chr6_+_31674639 | 0.24 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr1_-_54872059 | 0.24 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr4_+_166300084 | 0.24 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr9_+_125027127 | 0.22 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr3_+_52828805 | 0.21 |
ENST00000416872.2
ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr10_-_101945771 | 0.20 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr6_+_37225540 | 0.20 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr6_-_87804815 | 0.20 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr19_+_41281416 | 0.19 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr2_-_74779744 | 0.18 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr1_+_154966058 | 0.18 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr7_-_42276612 | 0.18 |
ENST00000395925.3
ENST00000437480.1 |
GLI3
|
GLI family zinc finger 3 |
| chr2_+_202316392 | 0.17 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr3_-_52719810 | 0.17 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr2_+_233271546 | 0.17 |
ENST00000295453.3
|
ALPPL2
|
alkaline phosphatase, placental-like 2 |
| chr9_-_79307096 | 0.17 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr4_+_169418255 | 0.16 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_132073111 | 0.16 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr18_+_54318616 | 0.15 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr19_+_41281282 | 0.14 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr2_-_74780176 | 0.13 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr11_-_46867780 | 0.13 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr19_+_41281060 | 0.13 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr1_+_109756523 | 0.12 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr20_-_31124186 | 0.12 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chrX_+_41548259 | 0.11 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr2_+_202937972 | 0.11 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr6_+_150070857 | 0.11 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chrX_+_41548220 | 0.11 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr5_-_81046904 | 0.11 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr11_+_129245796 | 0.11 |
ENST00000281437.4
|
BARX2
|
BARX homeobox 2 |
| chr10_+_94608218 | 0.11 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr12_+_62654155 | 0.11 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr5_-_81046922 | 0.10 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr4_-_46391805 | 0.10 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr8_-_99837856 | 0.10 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr1_+_66258846 | 0.10 |
ENST00000341517.4
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chrX_+_16804544 | 0.10 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr18_-_67872891 | 0.10 |
ENST00000454359.1
ENST00000437017.1 |
RTTN
|
rotatin |
| chr10_+_28822636 | 0.10 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr9_-_123639304 | 0.10 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr5_-_81046841 | 0.09 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr10_+_94608245 | 0.09 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr22_+_50946645 | 0.09 |
ENST00000420993.2
ENST00000395698.3 ENST00000395701.3 ENST00000523045.1 ENST00000299821.11 |
NCAPH2
|
non-SMC condensin II complex, subunit H2 |
| chr2_+_219524473 | 0.09 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr21_-_32931290 | 0.08 |
ENST00000286827.3
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr19_+_10828795 | 0.08 |
ENST00000389253.4
ENST00000355667.6 ENST00000408974.4 |
DNM2
|
dynamin 2 |
| chr3_-_52001448 | 0.08 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr5_+_125758865 | 0.08 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr14_-_21994525 | 0.08 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr16_+_88636789 | 0.08 |
ENST00000301011.5
ENST00000452588.2 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr20_-_3065362 | 0.08 |
ENST00000380293.3
|
AVP
|
arginine vasopressin |
| chr18_+_3449821 | 0.08 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_+_125758813 | 0.07 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr18_+_3449695 | 0.07 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrX_-_77225135 | 0.07 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr19_+_10828724 | 0.07 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr1_+_145549203 | 0.07 |
ENST00000355594.4
ENST00000544626.1 |
ANKRD35
|
ankyrin repeat domain 35 |
| chr11_+_45743858 | 0.07 |
ENST00000532307.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chr11_+_12132117 | 0.07 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr3_-_127541679 | 0.07 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr11_+_45743931 | 0.07 |
ENST00000530051.1
|
CTD-2210P24.1
|
CTD-2210P24.1 |
| chr18_+_54318566 | 0.07 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr5_-_127873896 | 0.07 |
ENST00000502468.1
|
FBN2
|
fibrillin 2 |
| chr4_+_140222609 | 0.06 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr1_-_179198702 | 0.06 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chrX_-_128788914 | 0.06 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr13_+_33160553 | 0.06 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr19_-_39735646 | 0.06 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr6_+_150070831 | 0.06 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr16_-_1275257 | 0.06 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr2_-_179672142 | 0.05 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr4_-_46391367 | 0.05 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr11_+_120207787 | 0.05 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_-_179112173 | 0.05 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr11_-_101000445 | 0.05 |
ENST00000534013.1
|
PGR
|
progesterone receptor |
| chr2_+_191208196 | 0.05 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr1_-_35658736 | 0.05 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr2_-_211179883 | 0.05 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr17_-_40337470 | 0.05 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr7_-_104909435 | 0.05 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr11_-_74660159 | 0.04 |
ENST00000527087.1
ENST00000321448.8 ENST00000340360.6 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr9_+_87285539 | 0.04 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr17_+_38474489 | 0.04 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr10_+_51565188 | 0.04 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr15_-_61521495 | 0.04 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr16_+_1756162 | 0.04 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr20_-_39946237 | 0.03 |
ENST00000441102.2
ENST00000559234.1 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr5_+_118407053 | 0.03 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr3_-_101395936 | 0.03 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr1_+_161736072 | 0.03 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr4_+_169418195 | 0.03 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr11_-_59383617 | 0.03 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr19_+_7733929 | 0.03 |
ENST00000221515.2
|
RETN
|
resistin |
| chr3_-_127541194 | 0.03 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr17_+_68071389 | 0.03 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_+_57050317 | 0.03 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr7_-_156803329 | 0.03 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr10_+_70661014 | 0.02 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr5_+_78985673 | 0.02 |
ENST00000446378.2
|
CMYA5
|
cardiomyopathy associated 5 |
| chr2_+_219081817 | 0.02 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr11_-_132813566 | 0.02 |
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like |
| chr16_+_57481349 | 0.02 |
ENST00000262507.6
ENST00000565964.1 |
COQ9
|
coenzyme Q9 |
| chr6_-_149867122 | 0.02 |
ENST00000253329.2
|
PPIL4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr7_-_137028498 | 0.02 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr17_+_68071458 | 0.02 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr7_-_137028534 | 0.02 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr6_+_108881012 | 0.02 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr17_-_4046257 | 0.02 |
ENST00000381638.2
|
ZZEF1
|
zinc finger, ZZ-type with EF-hand domain 1 |
| chr2_-_242842587 | 0.01 |
ENST00000404031.1
|
AC131097.4
|
Protein LOC285095 |
| chr6_-_114664180 | 0.01 |
ENST00000312719.5
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr11_+_74660278 | 0.01 |
ENST00000263672.6
ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr16_+_50776021 | 0.01 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr8_-_57123815 | 0.01 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr17_-_42452063 | 0.01 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr9_-_73477826 | 0.01 |
ENST00000396285.1
ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr3_+_52719936 | 0.01 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr22_+_30279144 | 0.01 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr12_-_91398796 | 0.01 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr18_-_54305658 | 0.00 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr19_+_36142147 | 0.00 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr2_-_202316260 | 0.00 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071947 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 0.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 3.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 1.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:1903977 | NMDA selective glutamate receptor signaling pathway(GO:0098989) positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 3.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.7 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |