Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
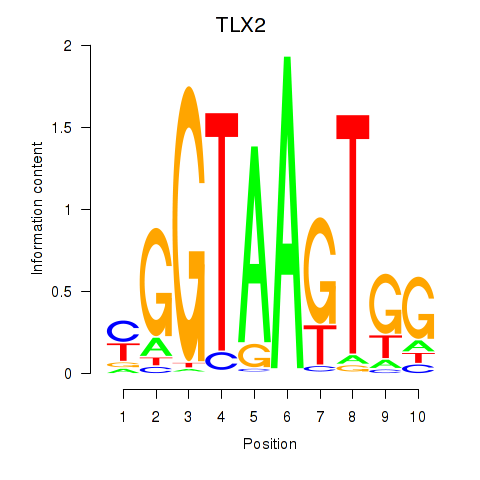
Results for TLX2
Z-value: 0.96
Transcription factors associated with TLX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX2
|
ENSG00000115297.9 | T cell leukemia homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TLX2 | hg19_v2_chr2_+_74741569_74741620 | -0.30 | 1.5e-01 | Click! |
Activity profile of TLX2 motif
Sorted Z-values of TLX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_57406368 | 5.74 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr1_-_169703203 | 5.37 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr6_-_31550192 | 3.76 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr6_-_143266297 | 3.61 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr5_+_131409476 | 2.98 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr19_+_676385 | 2.39 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr14_+_55034599 | 2.27 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr4_+_156680518 | 2.15 |
ENST00000513437.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr16_-_11681023 | 2.09 |
ENST00000570904.1
ENST00000574701.1 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr4_+_156680143 | 1.89 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr14_+_103589789 | 1.63 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr7_+_155090271 | 1.58 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr4_+_156680153 | 1.51 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr5_-_16916624 | 1.45 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr4_+_156588806 | 1.42 |
ENST00000513574.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr2_+_111878483 | 1.42 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr21_-_38639601 | 1.41 |
ENST00000539844.1
ENST00000476950.1 ENST00000399001.1 |
DSCR3
|
Down syndrome critical region gene 3 |
| chr16_-_11680759 | 1.39 |
ENST00000571459.1
ENST00000570798.1 ENST00000572255.1 ENST00000574763.1 ENST00000574703.1 ENST00000571277.1 ENST00000381810.3 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr12_-_89918522 | 1.36 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr2_+_102928009 | 1.36 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr8_+_22423219 | 1.35 |
ENST00000523965.1
ENST00000521554.1 |
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr12_-_57522813 | 1.32 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr17_+_66509019 | 1.23 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr8_-_91657740 | 1.17 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr15_-_50411412 | 1.16 |
ENST00000284509.6
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr10_+_35416090 | 1.15 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr10_+_35415719 | 1.12 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr16_-_11680791 | 1.10 |
ENST00000571976.1
ENST00000413364.2 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr21_+_43639211 | 1.06 |
ENST00000450121.1
ENST00000398449.3 ENST00000361802.2 |
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr3_-_195997410 | 0.99 |
ENST00000419333.1
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr1_+_214161272 | 0.98 |
ENST00000498508.2
ENST00000366958.4 |
PROX1
|
prospero homeobox 1 |
| chr16_-_67693846 | 0.94 |
ENST00000602850.1
|
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr14_-_92414055 | 0.94 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr10_+_35415978 | 0.93 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr20_-_1373682 | 0.93 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr3_-_182880541 | 0.91 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr10_+_35416223 | 0.90 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr3_-_149051194 | 0.90 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr3_+_30647994 | 0.87 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr6_+_106546808 | 0.84 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr6_+_37787704 | 0.84 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr3_-_107809816 | 0.83 |
ENST00000361309.5
ENST00000355354.7 |
CD47
|
CD47 molecule |
| chr5_-_150080472 | 0.81 |
ENST00000521464.1
ENST00000518917.1 ENST00000447771.2 ENST00000540000.1 ENST00000199814.4 |
RBM22
|
RNA binding motif protein 22 |
| chr10_-_60027642 | 0.80 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr4_+_1873100 | 0.80 |
ENST00000508803.1
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr10_-_123687431 | 0.80 |
ENST00000423243.1
|
ATE1
|
arginyltransferase 1 |
| chrX_-_45629661 | 0.79 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr2_-_225811747 | 0.79 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr7_+_112063192 | 0.77 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr20_+_44746939 | 0.76 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr8_-_105601134 | 0.74 |
ENST00000276654.5
ENST00000424843.2 |
LRP12
|
low density lipoprotein receptor-related protein 12 |
| chr2_-_204400113 | 0.73 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr1_-_161279749 | 0.73 |
ENST00000533357.1
ENST00000360451.6 ENST00000336559.4 |
MPZ
|
myelin protein zero |
| chr3_-_194373831 | 0.71 |
ENST00000437613.1
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr10_-_121632266 | 0.71 |
ENST00000360003.3
ENST00000369077.3 |
MCMBP
|
minichromosome maintenance complex binding protein |
| chr11_+_128563652 | 0.70 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_+_169923577 | 0.70 |
ENST00000432060.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr3_+_151986709 | 0.69 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr12_-_76953349 | 0.68 |
ENST00000551927.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr19_+_47104553 | 0.67 |
ENST00000598871.1
ENST00000594523.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chrX_+_70752945 | 0.66 |
ENST00000373701.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_-_22263790 | 0.64 |
ENST00000374695.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr2_+_150187020 | 0.64 |
ENST00000334166.4
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr2_+_10184302 | 0.61 |
ENST00000440320.1
ENST00000535335.1 |
KLF11
|
Kruppel-like factor 11 |
| chr1_-_68698197 | 0.58 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr4_+_169552748 | 0.58 |
ENST00000504519.1
ENST00000512127.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_50794730 | 0.57 |
ENST00000523389.1
ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr1_+_16174280 | 0.56 |
ENST00000375759.3
|
SPEN
|
spen family transcriptional repressor |
| chr22_-_22221658 | 0.56 |
ENST00000544786.1
|
MAPK1
|
mitogen-activated protein kinase 1 |
| chr19_-_12833361 | 0.55 |
ENST00000592287.1
|
TNPO2
|
transportin 2 |
| chr12_-_50677255 | 0.54 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr8_-_141728760 | 0.54 |
ENST00000430260.2
|
PTK2
|
protein tyrosine kinase 2 |
| chr20_-_47894569 | 0.54 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr12_-_51740463 | 0.54 |
ENST00000293636.1
|
CELA1
|
chymotrypsin-like elastase family, member 1 |
| chr11_-_57089671 | 0.54 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr15_-_38856836 | 0.53 |
ENST00000450598.2
ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr11_-_85780853 | 0.53 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_224033596 | 0.52 |
ENST00000391878.2
ENST00000343537.7 |
TP53BP2
|
tumor protein p53 binding protein, 2 |
| chr2_+_219433588 | 0.52 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr2_+_169923504 | 0.50 |
ENST00000357546.2
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr5_-_133968459 | 0.49 |
ENST00000505758.1
ENST00000439578.1 ENST00000502286.1 |
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr10_+_124030819 | 0.49 |
ENST00000260723.4
ENST00000368994.2 |
BTBD16
|
BTB (POZ) domain containing 16 |
| chr1_+_154229547 | 0.49 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr12_-_322504 | 0.48 |
ENST00000424061.2
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr9_+_116267536 | 0.48 |
ENST00000374136.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_+_107683644 | 0.48 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chr16_+_1876936 | 0.48 |
ENST00000382668.4
ENST00000382666.4 |
FAHD1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr10_+_35415851 | 0.47 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr2_+_10183651 | 0.47 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr2_+_219433281 | 0.47 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr10_-_43892279 | 0.47 |
ENST00000443950.2
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr10_-_50970322 | 0.46 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr2_-_175870085 | 0.46 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr20_+_2795626 | 0.46 |
ENST00000603872.1
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr10_-_123687943 | 0.46 |
ENST00000540606.1
ENST00000455628.1 |
ATE1
|
arginyltransferase 1 |
| chr20_+_44746885 | 0.46 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chrX_-_118826784 | 0.46 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr18_+_55862622 | 0.46 |
ENST00000456173.2
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr22_+_42229100 | 0.45 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr9_+_116225999 | 0.44 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr15_+_89631647 | 0.44 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr12_+_9144626 | 0.43 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr20_-_42839378 | 0.43 |
ENST00000255174.2
|
OSER1
|
oxidative stress responsive serine-rich 1 |
| chr12_-_76953513 | 0.43 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr20_-_61557821 | 0.43 |
ENST00000354665.4
ENST00000370368.1 ENST00000395343.1 ENST00000395340.1 |
DIDO1
|
death inducer-obliterator 1 |
| chr10_-_50970382 | 0.43 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr1_+_24286287 | 0.42 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr10_+_76871229 | 0.42 |
ENST00000372690.3
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr17_+_42925270 | 0.41 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr6_-_43496605 | 0.40 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr12_-_71031185 | 0.40 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr10_-_123687399 | 0.39 |
ENST00000543447.1
|
ATE1
|
arginyltransferase 1 |
| chr9_+_139847347 | 0.39 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr19_+_36236491 | 0.39 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr12_-_48119301 | 0.39 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr16_+_31404624 | 0.39 |
ENST00000389202.2
|
ITGAD
|
integrin, alpha D |
| chr21_-_16437126 | 0.39 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr19_-_4517613 | 0.39 |
ENST00000301286.3
|
PLIN4
|
perilipin 4 |
| chr5_+_133984462 | 0.38 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr12_+_69864129 | 0.38 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr21_-_16437255 | 0.38 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr14_-_21737551 | 0.38 |
ENST00000554891.1
ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr19_+_36236514 | 0.38 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr14_+_70233810 | 0.37 |
ENST00000394366.2
ENST00000553548.1 ENST00000553369.1 ENST00000557154.1 ENST00000451983.2 ENST00000553635.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr17_-_76899275 | 0.37 |
ENST00000322630.2
ENST00000586713.1 |
DDC8
|
Protein DDC8 homolog |
| chr2_-_175869936 | 0.36 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chrX_-_77395186 | 0.36 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr8_-_145018905 | 0.36 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr1_-_17304771 | 0.36 |
ENST00000375534.3
|
MFAP2
|
microfibrillar-associated protein 2 |
| chrX_+_70752917 | 0.36 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr12_-_48119340 | 0.36 |
ENST00000229003.3
|
ENDOU
|
endonuclease, polyU-specific |
| chr1_+_160160283 | 0.35 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr20_-_1373726 | 0.35 |
ENST00000400137.4
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr8_+_91952750 | 0.34 |
ENST00000521366.1
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1 |
| chr1_-_217262969 | 0.34 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr12_+_113229737 | 0.34 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr12_-_113658892 | 0.34 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr1_+_160160346 | 0.34 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr7_+_45197383 | 0.34 |
ENST00000242249.4
ENST00000496212.1 ENST00000481345.1 |
RAMP3
|
receptor (G protein-coupled) activity modifying protein 3 |
| chr19_+_47104493 | 0.33 |
ENST00000291295.9
ENST00000597743.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr12_+_54519842 | 0.33 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr5_+_133451254 | 0.33 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr1_-_204135450 | 0.33 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr5_-_147162078 | 0.33 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr8_+_62737875 | 0.33 |
ENST00000523042.1
ENST00000518593.1 ENST00000519452.1 ENST00000519967.1 |
RP11-705O24.1
|
RP11-705O24.1 |
| chr15_+_89631381 | 0.32 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chrX_+_38420783 | 0.32 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr11_-_3818688 | 0.32 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr1_-_31769656 | 0.31 |
ENST00000446633.2
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5) |
| chr12_+_65672702 | 0.31 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr12_+_57522439 | 0.30 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr12_+_50478977 | 0.30 |
ENST00000381513.4
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr1_-_17766198 | 0.30 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr17_+_66508537 | 0.30 |
ENST00000392711.1
ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr20_+_61569463 | 0.30 |
ENST00000266069.3
|
GID8
|
GID complex subunit 8 |
| chr1_-_217262933 | 0.30 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr19_-_1876156 | 0.30 |
ENST00000565797.1
|
CTB-31O20.2
|
CTB-31O20.2 |
| chr6_+_84563295 | 0.30 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr18_+_29077990 | 0.30 |
ENST00000261590.8
|
DSG2
|
desmoglein 2 |
| chr6_+_111303218 | 0.30 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr2_+_242255275 | 0.30 |
ENST00000391971.2
|
SEPT2
|
septin 2 |
| chr6_+_74072375 | 0.30 |
ENST00000370367.3
|
KHDC3L
|
KH domain containing 3-like, subcortical maternal complex member |
| chr14_-_21737610 | 0.29 |
ENST00000320084.7
ENST00000449098.1 ENST00000336053.6 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr2_-_204400013 | 0.29 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr5_-_147162263 | 0.29 |
ENST00000333010.6
ENST00000265272.5 |
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr3_+_46412345 | 0.29 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr1_+_207627575 | 0.28 |
ENST00000367058.3
ENST00000367057.3 ENST00000367059.3 |
CR2
|
complement component (3d/Epstein Barr virus) receptor 2 |
| chr19_+_39421556 | 0.28 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr11_+_64058758 | 0.28 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr16_-_70835034 | 0.28 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr8_-_131455835 | 0.28 |
ENST00000518721.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr15_-_34502197 | 0.27 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr2_+_191045562 | 0.27 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chrX_-_73512411 | 0.27 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr10_-_27529486 | 0.27 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr1_-_153517473 | 0.27 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr3_+_121796697 | 0.27 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chrX_-_72095622 | 0.27 |
ENST00000290273.5
|
DMRTC1
|
DMRT-like family C1 |
| chr7_+_54610086 | 0.27 |
ENST00000404951.1
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chrX_+_38420623 | 0.27 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr6_+_37787458 | 0.26 |
ENST00000373391.2
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr17_+_7211280 | 0.26 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr2_-_228497888 | 0.26 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chrX_-_38186811 | 0.26 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr19_-_52150053 | 0.26 |
ENST00000599649.1
ENST00000429354.3 ENST00000360844.6 ENST00000222107.4 |
SIGLEC5
SIGLEC14
SIGLEC5
|
SIGLEC5 sialic acid binding Ig-like lectin 14 sialic acid binding Ig-like lectin 5 |
| chr3_-_112693865 | 0.26 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr12_-_76953453 | 0.26 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr10_-_124768300 | 0.26 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr19_+_10400615 | 0.26 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr16_+_81478775 | 0.26 |
ENST00000537098.3
|
CMIP
|
c-Maf inducing protein |
| chr1_+_207627697 | 0.26 |
ENST00000458541.2
|
CR2
|
complement component (3d/Epstein Barr virus) receptor 2 |
| chr7_+_130794846 | 0.26 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr14_-_57272366 | 0.26 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr1_-_183622442 | 0.26 |
ENST00000308641.4
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr1_+_214161854 | 0.26 |
ENST00000435016.1
|
PROX1
|
prospero homeobox 1 |
| chr15_-_40401062 | 0.25 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr12_+_71833756 | 0.25 |
ENST00000536515.1
ENST00000540815.2 |
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr3_+_122283064 | 0.25 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr1_+_205473720 | 0.25 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr20_-_1373606 | 0.25 |
ENST00000381715.1
ENST00000439640.2 ENST00000381719.3 |
FKBP1A
|
FK506 binding protein 1A, 12kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.7 | 1.4 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.7 | 5.5 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.6 | 3.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) negative regulation of cytolysis(GO:0045918) |
| 0.5 | 3.8 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.5 | 1.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.4 | 1.7 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.4 | 1.2 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.4 | 0.4 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.3 | 1.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.3 | 0.9 | GO:0060381 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.3 | 0.9 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.3 | 1.1 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.3 | 0.8 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.3 | 1.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.2 | 1.2 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.2 | 5.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 2.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.4 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.2 | 0.7 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 1.1 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 0.2 | 1.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 1.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 0.5 | GO:0045404 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 0.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.4 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 4.9 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.5 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 1.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.5 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 1.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.3 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.1 | 0.5 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 1.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 1.0 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 5.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.5 | GO:1902962 | positive regulation of beta-amyloid clearance(GO:1900223) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 0.3 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.7 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.5 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.3 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.1 | 0.2 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.3 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.1 | 0.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.8 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.8 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 1.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 2.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0051142 | positive regulation of activation of Janus kinase activity(GO:0010536) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.3 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.3 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.8 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 1.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.3 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.2 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.5 | 2.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 1.5 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.3 | 0.9 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.3 | 0.8 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.3 | 7.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 0.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 0.5 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.2 | 1.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 1.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 1.1 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 1.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 6.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 3.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 2.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 2.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.4 | 1.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 1.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.3 | 5.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 4.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 7.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 1.0 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.2 | 0.9 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 3.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 0.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.8 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.2 | 1.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.4 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 5.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.6 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 1.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.5 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 4.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 2.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 2.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 3.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 7.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 5.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 2.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 5.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 1.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 2.1 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 1.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |