|
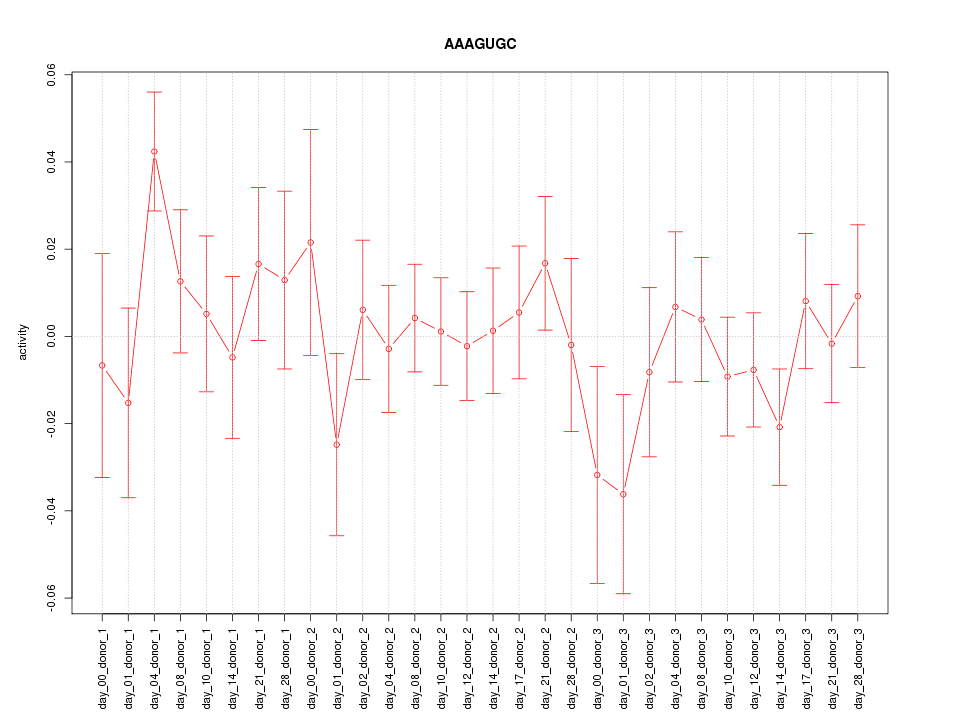
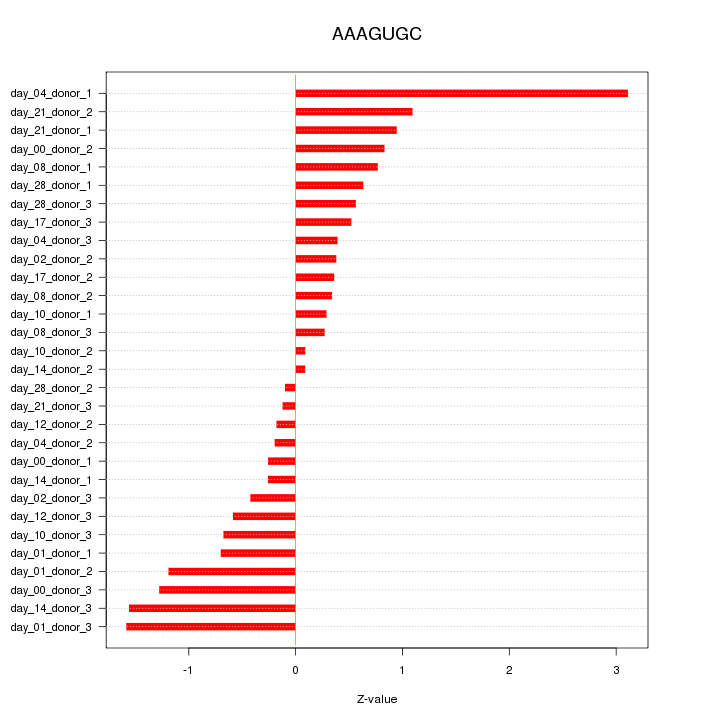
chr9_-_4152182
|
2.303
|
NM_152629
|
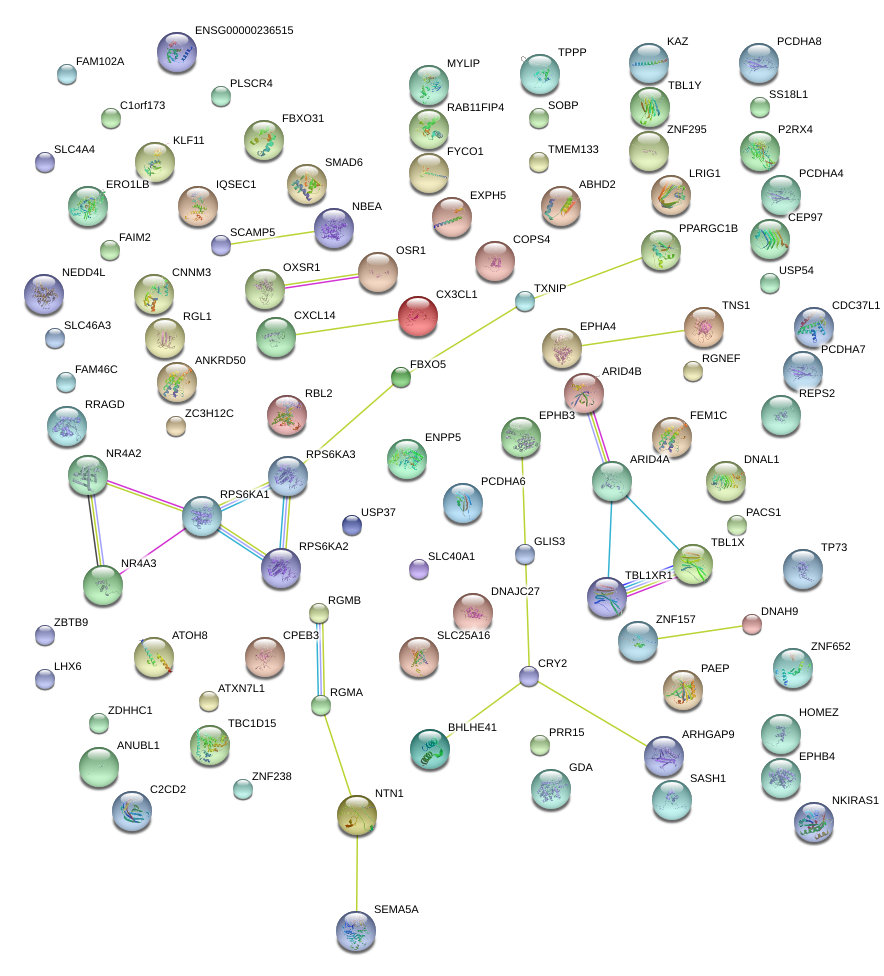
GLIS3
|
GLIS family zinc finger 3
|
|
chr9_-_4300028
|
2.267
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr13_+_36050885
|
2.118
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr13_+_35516390
|
1.792
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr1_+_118148506
|
1.698
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr6_-_90121966
|
1.329
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr9_-_124984018
|
1.294
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr11_+_100862719
|
1.226
|
NM_032021
|
TMEM133
|
transmembrane protein 133
|
|
chr2_-_19558371
|
1.166
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr9_-_124990949
|
1.163
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chrX_+_16964730
|
1.159
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr4_+_72052354
|
1.147
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr9_-_124990706
|
1.139
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chrX_+_9431314
|
1.093
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr3_-_66550844
|
1.075
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr3_-_145968958
|
1.068
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr2_-_190445521
|
1.036
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr1_+_244214552
|
1.011
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr2_-_222436996
|
1.003
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr1_+_183605207
|
0.979
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr9_+_74729510
|
0.972
|
NM_001242507
|
GDA
|
guanine deaminase
|
|
chr2_+_85980600
|
0.971
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr16_+_57406398
|
0.966
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr5_+_149151502
|
0.963
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr11_+_109964086
|
0.955
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr6_-_167275657
|
0.925
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr13_-_29292961
|
0.923
|
NM_001135919
NM_181785
|
SLC46A3
|
solute carrier family 46, member 3
|
|
chr1_+_244216506
|
0.914
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr2_-_157189040
|
0.905
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr5_+_149109814
|
0.902
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr6_-_167040700
|
0.890
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chrX_+_9432980
|
0.884
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr5_-_134914968
|
0.882
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr1_-_75139421
|
0.872
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr21_-_43373938
|
0.867
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr6_-_46138584
|
0.843
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr5_-_9546157
|
0.800
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr11_-_108464344
|
0.795
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr9_-_124989755
|
0.788
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr12_+_72233486
|
0.773
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr4_+_72204769
|
0.773
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr2_+_97481977
|
0.752
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr7_+_29603388
|
0.702
|
NM_175887
|
PRR15
|
proline rich 15
|
|
chr15_-_93616891
|
0.698
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr6_+_33422319
|
0.677
|
NM_152735
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
|
chr11_+_45868668
|
0.671
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr9_+_4679557
|
0.667
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr12_-_89918531
|
0.665
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr1_+_3569098
|
0.663
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr11_+_45868956
|
0.663
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr2_-_25194772
|
0.649
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr21_-_43346780
|
0.649
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr15_-_93632161
|
0.646
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr18_+_55888750
|
0.642
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_+_107811272
|
0.641
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr9_-_130742803
|
0.640
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr5_-_693413
|
0.631
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr15_-_93617091
|
0.629
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr7_-_105319608
|
0.627
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr9_+_74764266
|
0.626
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr6_+_148663728
|
0.624
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr15_-_93616358
|
0.623
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr3_-_13008959
|
0.622
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr15_+_67000929
|
0.618
|
NM_001142861
|
SMAD6
|
SMAD family member 6
|
|
chr1_+_3607232
|
0.618
|
NM_001126240
NM_001126241
NM_001126242
NM_001204189
NM_001204190
NM_001204191
|
TP73
|
tumor protein p73
|
|
chr3_+_101443436
|
0.611
|
NM_024548
|
CEP97
|
centrosomal protein 97kDa
|
|
chr15_-_93631751
|
0.611
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr3_-_46037299
|
0.610
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr5_+_140254930
|
0.600
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr1_+_145438437
|
0.596
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr12_-_26277807
|
0.589
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr5_+_140213968
|
0.588
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr1_+_3614609
|
0.585
|
NM_001204192
|
TP73
|
tumor protein p73
|
|
chr15_+_75287829
|
0.581
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr17_+_29718641
|
0.581
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr3_-_23958500
|
0.580
|
NM_020345
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1
|
|
chr21_-_43430448
|
0.579
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr17_+_8924822
|
0.576
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr2_-_219432955
|
0.574
|
NM_020935
|
USP37
|
ubiquitin specific peptidase 37
|
|
chr16_+_53468344
|
0.572
|
NM_005611
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr5_+_140220811
|
0.569
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr16_-_67450149
|
0.559
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr10_-_46168156
|
0.558
|
NM_001128324
NM_174890
|
ZFAND4
|
zinc finger, AN1-type domain 4
|
|
chr5_+_140186658
|
0.551
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr2_+_10184371
|
0.549
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr2_-_218808705
|
0.548
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr4_-_125633716
|
0.540
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr17_-_47439326
|
0.540
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr10_-_94050799
|
0.540
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr2_+_10183630
|
0.535
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr12_+_121647659
|
0.533
|
NM_002560
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4
|
|
chr20_+_60718473
|
0.532
|
NM_198935
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr10_-_94003002
|
0.525
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr5_+_72921980
|
0.521
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr15_+_89631380
|
0.513
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr6_+_16129285
|
0.506
|
NM_013262
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr5_+_98104998
|
0.505
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr14_+_74111577
|
0.503
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr1_-_236445294
|
0.501
|
NM_019891
|
ERO1LB
|
ERO1-like beta (S. cerevisiae)
|
|
chr5_+_140247767
|
0.499
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr5_-_114880536
|
0.496
|
NM_020177
|
FEM1C
|
fem-1 homolog c (C. elegans)
|
|
chr12_-_50297575
|
0.495
|
NM_012306
|
FAIM2
|
Fas apoptotic inhibitory molecule 2
|
|
chr14_+_58765209
|
0.492
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr16_-_87417309
|
0.485
|
NM_024735
|
FBXO31
|
F-box protein 31
|
|
chr18_+_55714457
|
0.483
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr5_+_140207562
|
0.472
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr17_-_38083762
|
0.470
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr1_+_67218104
|
0.467
|
NM_152665
|
TCTEX1D1
|
Tctex1 domain containing 1
|
|
chr22_+_30279064
|
0.467
|
NM_021090
NM_153050
NM_153051
|
MTMR3
|
myotubularin related protein 3
|
|
chr16_+_69599868
|
0.460
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr18_+_55711388
|
0.458
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_-_24045302
|
0.455
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr3_-_13114547
|
0.452
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr4_-_53525316
|
0.446
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr11_-_108093328
|
0.444
|
NM_002519
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chr21_-_36421586
|
0.442
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr19_+_38397841
|
0.441
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr1_-_157015161
|
0.438
|
NM_014784
NM_198236
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11
|
|
chr5_-_122759003
|
0.438
|
NM_001166226
NM_153223
|
CEP120
|
centrosomal protein 120kDa
|
|
chr14_-_89020888
|
0.438
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr3_+_140950671
|
0.435
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr6_-_137113624
|
0.434
|
NM_005923
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr17_+_41132581
|
0.431
|
NM_173079
|
RUNDC1
|
RUN domain containing 1
|
|
chr18_+_9137560
|
0.431
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr10_+_120789227
|
0.430
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr14_-_38064440
|
0.422
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr18_+_67956136
|
0.422
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr17_+_4981721
|
0.422
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr15_+_86220267
|
0.420
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr9_-_112260529
|
0.419
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr4_-_85887543
|
0.419
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr2_-_197036301
|
0.418
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr2_-_240322620
|
0.417
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr8_+_107460151
|
0.416
|
NM_001198532
|
OXR1
|
oxidation resistance 1
|
|
chr9_-_112213663
|
0.416
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr5_+_142150291
|
0.414
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chrX_-_53310748
|
0.414
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr16_+_14164879
|
0.414
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr11_-_113746205
|
0.408
|
NM_020886
|
USP28
|
ubiquitin specific peptidase 28
|
|
chr11_-_6494985
|
0.407
|
NM_001248007
NM_006458
NM_033278
|
TRIM3
|
tripartite motif containing 3
|
|
chr12_+_131438451
|
0.405
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr4_+_7045142
|
0.405
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr15_+_85923739
|
0.404
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr10_-_61469322
|
0.404
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr14_-_92506284
|
0.402
|
NM_004239
|
TRIP11
|
thyroid hormone receptor interactor 11
|
|
chr8_+_107669895
|
0.399
|
NM_181354
|
OXR1
|
oxidation resistance 1
|
|
chr20_-_47444404
|
0.399
|
NM_020820
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1
|
|
chr16_+_1664640
|
0.398
|
NM_020825
|
CRAMP1L
|
Crm, cramped-like (Drosophila)
|
|
chr3_+_20081523
|
0.395
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr17_+_57408907
|
0.394
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chrY_+_2803409
|
0.388
|
NM_001145275
NM_003411
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr14_+_60712469
|
0.387
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr5_+_140165875
|
0.385
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr18_-_30050394
|
0.383
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr18_-_72921280
|
0.382
|
NM_175907
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2
|
|
chr4_-_89744154
|
0.381
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr8_+_86099909
|
0.380
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr2_+_179345194
|
0.380
|
NM_019091
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3
|
|
chr18_+_10454620
|
0.376
|
NM_153000
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr18_+_55862577
|
0.375
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrY_+_2803111
|
0.374
|
NM_001145276
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr6_-_47277646
|
0.372
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr19_+_50380681
|
0.369
|
NM_001168222
NM_024682
|
TBC1D17
|
TBC1 domain family, member 17
|
|
chr22_-_22089975
|
0.368
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr8_+_107738239
|
0.365
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr12_-_65153189
|
0.364
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr4_-_53522756
|
0.361
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr9_-_112191105
|
0.360
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr18_+_55816546
|
0.359
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_+_119384490
|
0.358
|
NM_006777
NM_001184742
|
ZBTB33
|
zinc finger and BTB domain containing 33
|
|
chr8_+_86089618
|
0.357
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr1_-_160232286
|
0.354
|
NM_015726
|
DCAF8
|
DDB1 and CUL4 associated factor 8
|
|
chr6_-_79787939
|
0.351
|
NM_017934
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr15_-_52483564
|
0.349
|
NM_016194
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr7_+_30174343
|
0.347
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr11_-_73309090
|
0.347
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr19_+_18208596
|
0.337
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr21_+_45432202
|
0.337
|
NM_003274
|
TRAPPC10
|
trafficking protein particle complex 10
|
|
chr11_+_48002063
|
0.336
|
NM_001098503
NM_002843
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr12_+_56414688
|
0.335
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr2_-_96931744
|
0.334
|
NM_001193304
NM_017849
|
TMEM127
|
transmembrane protein 127
|
|
chr10_+_93558139
|
0.334
|
NM_025235
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr1_-_151798552
|
0.332
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chrX_-_10851808
|
0.330
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_+_33722158
|
0.330
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chrX_-_108976509
|
0.330
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr12_+_56473808
|
0.326
|
NM_001982
NM_001005915
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr8_-_82754427
|
0.322
|
NM_022133
NM_152836
NM_152837
|
SNX16
|
sorting nexin 16
|
|
chr7_-_2354050
|
0.321
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr12_-_89920038
|
0.320
|
NM_001199781
NM_001199782
NM_172240
|
POC1B-GALNT4
POC1B
|
POC1B-GALNT4 readthrough
POC1 centriolar protein homolog B (Chlamydomonas)
|
|
chr15_+_44719578
|
0.319
|
NM_016396
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr21_+_44394634
|
0.319
|
NM_004571
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr12_-_117628112
|
0.315
|
NM_015002
NM_033624
|
FBXO21
|
F-box protein 21
|
|
chr5_-_133968463
|
0.310
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr7_+_130794854
|
0.307
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr17_-_40897025
|
0.306
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|
|
chr11_-_130184459
|
0.303
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr20_+_51801821
|
0.302
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr15_-_52472075
|
0.301
|
NM_006578
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr16_-_1429612
|
0.298
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|