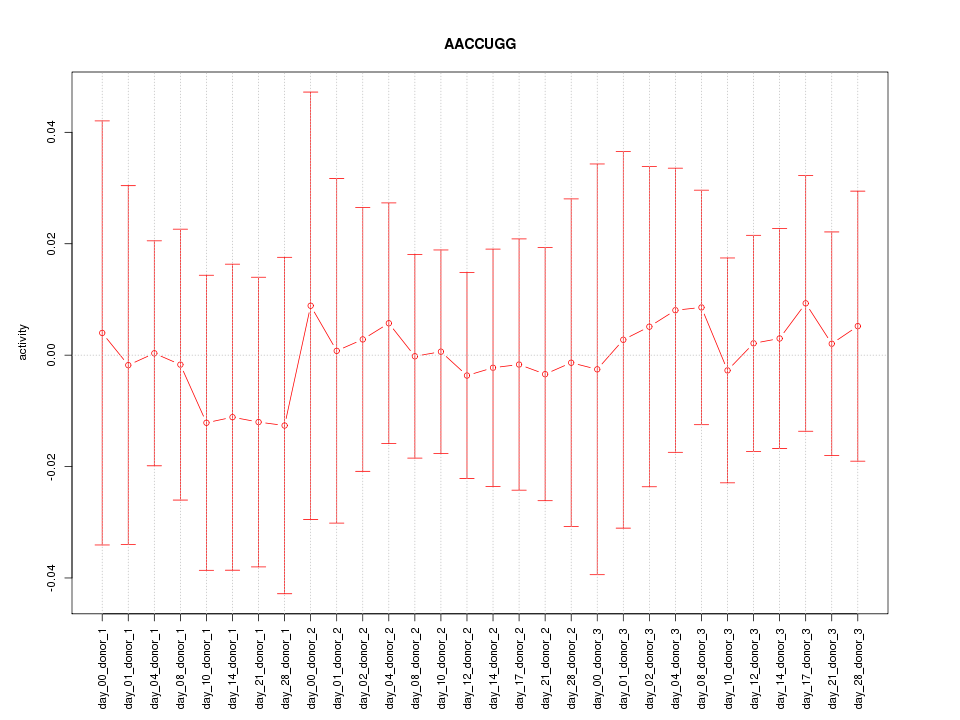
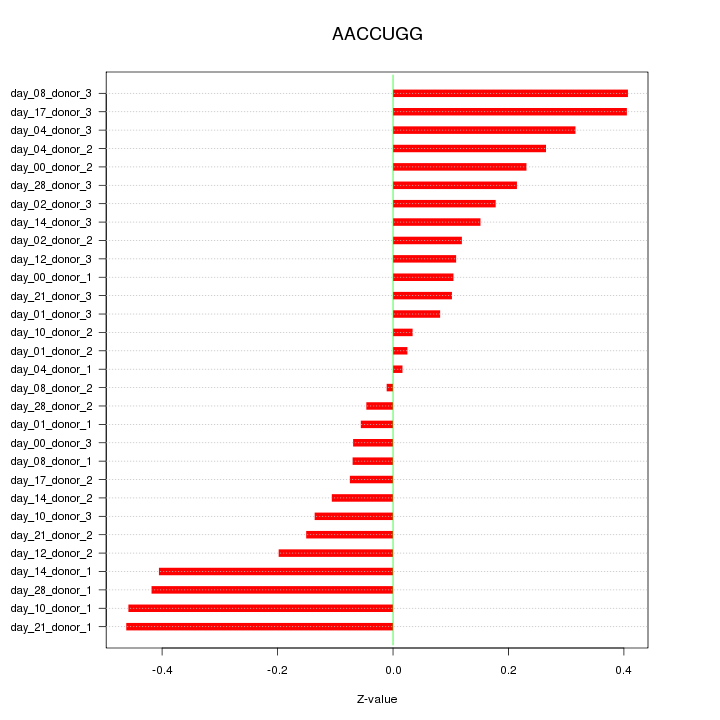
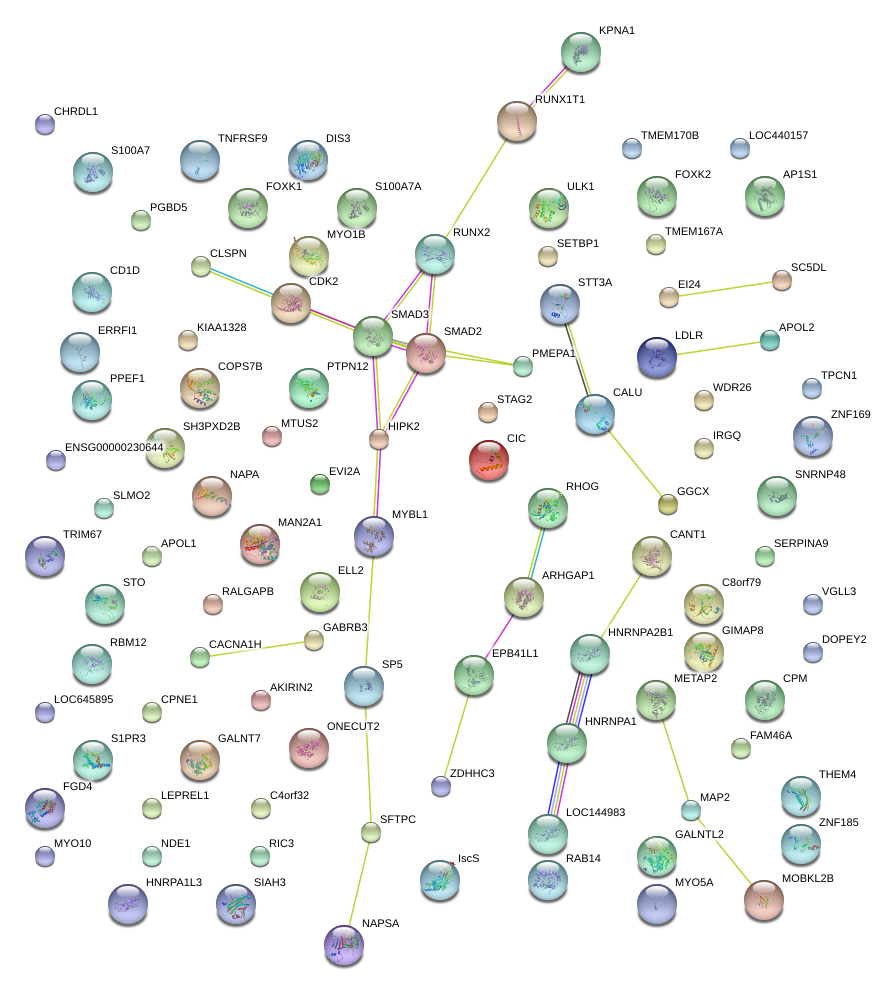
|
chr12_-_69357019
|
0.324
|
NM_001874
|
CPM
|
carboxypeptidase M
|
|
chr12_-_69326946
|
0.308
|
NM_001005502
NM_198320
|
CPM
|
carboxypeptidase M
|
|
chr1_-_230513390
|
0.201
|
NM_024554
|
PGBD5
|
piggyBac transposable element derived 5
|
|
chr7_+_100797685
|
0.180
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr7_+_77167351
|
0.157
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr7_+_77166750
|
0.154
|
NM_002835
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr11_-_8190535
|
0.154
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr1_+_153388999
|
0.128
|
NM_176823
|
S100A7A
|
S100 calcium binding protein A7A
|
|
chr6_-_88411984
|
0.107
|
NM_018064
|
AKIRIN2
|
akirin 2
|
|
chr8_-_93111915
|
0.098
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_110038992
|
0.094
|
NM_145234
|
CHRDL1
|
chordin-like 1
|
|
chr7_-_139477437
|
0.088
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr15_-_26874236
|
0.087
|
NM_001191321
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr8_-_93029864
|
0.084
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_110039285
|
0.083
|
NM_001143981
NM_001143982
NM_001143983
|
CHRDL1
|
chordin-like 1
|
|
chr22_+_36649116
|
0.083
|
NM_001136540
NM_001136541
NM_003661
NM_145343
|
APOL1
|
apolipoprotein L, 1
|
|
chr21_+_37536838
|
0.077
|
NM_005128
|
DOPEY2
|
dopey family member 2
|
|
chr20_-_56286540
|
0.071
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr8_-_93107881
|
0.069
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_-_26961911
|
0.069
|
NM_001191320
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr2_+_210444364
|
0.066
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr20_-_56284945
|
0.065
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr8_-_93115453
|
0.065
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr19_+_11200037
|
0.064
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr12_+_54674478
|
0.064
|
NM_002136
NM_031157
|
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1
|
|
chrX_+_152110214
|
0.063
|
NM_001178115
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr8_-_93107442
|
0.062
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_+_152082985
|
0.061
|
NM_001178106
NM_001178107
NM_001178108
NM_001178109
NM_001178110
NM_007150
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr2_+_171571800
|
0.061
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr1_-_8086392
|
0.061
|
NM_018948
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr8_-_93075190
|
0.060
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_+_123094377
|
0.060
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr2_+_210288668
|
0.057
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr3_-_189840225
|
0.057
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr5_+_109025135
|
0.057
|
NM_002372
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chrX_+_152086408
|
0.057
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr20_-_56285576
|
0.056
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr11_+_125462736
|
0.056
|
NM_152713
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr4_+_113066552
|
0.055
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr3_-_87040246
|
0.054
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr15_-_27018917
|
0.053
|
NM_021912
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr18_+_42260774
|
0.053
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr9_-_123964337
|
0.053
|
NM_016322
|
RAB14
|
RAB14, member RAS oncogene family
|
|
chr22_-_36635666
|
0.052
|
NM_030882
|
APOL2
|
apolipoprotein L, 2
|
|
chr22_-_36635999
|
0.051
|
NM_145637
|
APOL2
|
apolipoprotein L, 2
|
|
chr12_+_113659216
|
0.050
|
NM_001143819
NM_017901
|
TPCN1
|
two pore segment channel 1
|
|
chr6_+_11538486
|
0.045
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr9_-_27529434
|
0.044
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr3_-_45017600
|
0.042
|
NM_001135179
NM_016598
|
ZDHHC3
|
zinc finger, DHHC-type containing 3
|
|
chr20_+_34700257
|
0.042
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr4_+_174089889
|
0.042
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr11_+_121163501
|
0.041
|
NM_001024956
|
SC5DL
|
sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like
|
|
chr14_-_94942513
|
0.041
|
NM_001042518
NM_175739
|
SERPINA9
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9
|
|
chr7_+_128379345
|
0.040
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr2_+_192109879
|
0.040
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr11_+_121163343
|
0.039
|
NM_006918
|
SC5DL
|
sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like
|
|
chr15_-_52821004
|
0.039
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr20_-_57617836
|
0.036
|
NM_016045
|
SLMO2
|
slowmo homolog 2 (Drosophila)
|
|
chrX_+_18709044
|
0.036
|
NM_006240
NM_152224
NM_152226
|
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1
|
|
chr3_-_189838907
|
0.036
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr5_-_82373253
|
0.035
|
NM_174909
|
TMEM167A
|
transmembrane protein 167A
|
|
chr11_-_3862183
|
0.035
|
NM_001665
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr19_-_48018317
|
0.035
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr15_+_67358194
|
0.032
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr2_+_232651059
|
0.032
|
NM_022730
|
COPS7B
|
COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis)
|
|
chr17_-_29648631
|
0.031
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr5_-_16936371
|
0.029
|
NM_012334
|
MYO10
|
myosin X
|
|
chr16_+_1203240
|
0.028
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr19_-_44100127
|
0.028
|
NM_001007561
|
IRGQ
|
immunity-related GTPase family, Q
|
|
chr13_-_73356070
|
0.027
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr15_+_67418053
|
0.027
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr5_-_171881421
|
0.026
|
NM_001017995
|
SH3PXD2B
|
SH3 and PX domains 2B
|
|
chr1_-_36235422
|
0.026
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr13_+_30002776
|
0.025
|
NM_015233
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr15_-_27018216
|
0.024
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr15_+_67430358
|
0.023
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr6_+_45389818
|
0.023
|
NM_004348
|
RUNX2
|
runt-related transcription factor 2
|
|
chr12_+_132379262
|
0.021
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr11_-_46722115
|
0.021
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr5_+_176560056
|
0.020
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr15_+_67458492
|
0.020
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr6_-_82462374
|
0.020
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr8_-_67525426
|
0.019
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr1_-_8003224
|
0.019
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr6_+_7590405
|
0.018
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr19_+_42788605
|
0.018
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr1_+_158149736
|
0.018
|
NM_001766
|
CD1D
|
CD1d molecule
|
|
chr6_+_45296053
|
0.018
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr5_+_176560627
|
0.018
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr5_-_95297597
|
0.018
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr7_+_150147936
|
0.017
|
NM_175571
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr7_-_26240366
|
0.017
|
NM_002137
NM_031243
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr1_-_151882360
|
0.017
|
NM_053055
|
THEM4
|
thioesterase superfamily member 4
|
|
chr8_+_12803150
|
0.017
|
NM_020844
|
KIAA1456
|
KIAA1456
|
|
chr12_+_32654976
|
0.017
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr13_-_46425756
|
0.016
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr1_-_224622000
|
0.016
|
NM_001115113
NM_025160
|
WDR26
|
WD repeat domain 26
|
|
chr2_-_85788644
|
0.016
|
NM_000821
NM_001142269
|
GGCX
|
gamma-glutamyl carboxylase
|
|
chr18_+_55102777
|
0.016
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr7_+_4721722
|
0.015
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr17_-_77005822
|
0.015
|
NM_001159772
NM_001159773
NM_138793
|
CANT1
|
calcium activated nucleotidase 1
|
|
chr8_+_12869772
|
0.015
|
NM_001099677
|
KIAA1456
|
KIAA1456
|
|
chr16_+_15737123
|
0.015
|
NM_001143979
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr1_+_231298673
|
0.014
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr3_-_122233781
|
0.014
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|
|
chr9_+_91606349
|
0.014
|
NM_005226
|
S1PR3
|
sphingosine-1-phosphate receptor 3
|
|
chr20_-_34252861
|
0.014
|
NM_001198838
NM_001198840
NM_006047
NM_152838
NM_001198863
NM_152925
NM_152926
NM_152927
NM_152928
|
RBM12
CPNE1
|
RNA binding motif protein 12
copine I
|
|
chr18_+_34409058
|
0.014
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr12_+_56360547
|
0.013
|
NM_001798
NM_052827
|
CDK2
|
cyclin-dependent kinase 2
|
|
chr20_+_37101457
|
0.013
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr11_+_125439294
|
0.013
|
NM_001007277
NM_004879
|
EI24
|
etoposide induced 2.4 mRNA
|
|
chr3_+_114012832
|
0.013
|
NM_173799
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains
|
|
chr7_+_128828712
|
0.013
|
NM_005631
|
SMO
|
smoothened, frizzled family receptor
|
|
chr1_+_145096391
|
0.013
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr4_+_56212381
|
0.013
|
NM_024592
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr17_-_48785016
|
0.012
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr19_-_2456954
|
0.012
|
NM_032737
|
LMNB2
|
lamin B2
|
|
chr19_+_56915693
|
0.012
|
NM_152478
NM_001159861
|
ZNF583
|
zinc finger protein 583
|
|
chr4_+_1902352
|
0.012
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr1_+_145524989
|
0.012
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr12_+_2162388
|
0.011
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr19_+_56915382
|
0.011
|
NM_001159860
|
ZNF583
|
zinc finger protein 583
|
|
chr12_-_21487831
|
0.011
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr1_-_150849211
|
0.010
|
NM_001197325
NM_001668
NM_178427
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr17_-_8022134
|
0.010
|
NM_001165960
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr10_-_52645430
|
0.010
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr11_+_18720342
|
0.010
|
NM_153347
|
TMEM86A
|
transmembrane protein 86A
|
|
chr18_+_55019720
|
0.010
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr4_+_699529
|
0.010
|
NM_006315
|
PCGF3
|
polycomb group ring finger 3
|
|
chr1_-_226187055
|
0.010
|
NM_152608
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chrX_-_65835845
|
0.010
|
NM_001242310
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr2_+_97001463
|
0.009
|
NM_015341
|
NCAPH
|
non-SMC condensin I complex, subunit H
|
|
chr20_+_34742656
|
0.009
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr1_-_173572179
|
0.009
|
NM_178527
|
SLC9A11
|
solute carrier family 9, member 11
|
|
chr18_-_33647336
|
0.009
|
NM_018170
|
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A
|
|
chr1_-_201475966
|
0.009
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr1_-_201476248
|
0.009
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chrX_-_65859107
|
0.009
|
NM_001199687
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr12_-_56615484
|
0.009
|
NM_001242826
|
RNF41
|
ring finger protein 41
|
|
chr12_-_56615679
|
0.009
|
NM_005785
NM_194358
NM_194359
|
RNF41
|
ring finger protein 41
|
|
chr22_-_33454328
|
0.009
|
NM_001135774
|
SYN3
|
synapsin III
|
|
chr13_+_50202434
|
0.009
|
NM_138450
|
ARL11
|
ADP-ribosylation factor-like 11
|
|
chr3_+_196295708
|
0.008
|
NM_001105573
|
FBXO45
|
F-box protein 45
|
|
chr16_+_15744082
|
0.008
|
NM_017668
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr6_+_34759743
|
0.008
|
NM_017754
|
UHRF1BP1
|
UHRF1 binding protein 1
|
|
chr2_+_48757292
|
0.008
|
NM_001198595
NM_006873
|
STON1
|
stonin 1
|
|
chrX_-_65858882
|
0.008
|
NM_021783
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr8_-_101315457
|
0.007
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr12_+_1929432
|
0.007
|
NM_001039029
NM_001163925
NM_001163926
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr1_-_201465700
|
0.007
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr17_-_1303548
|
0.007
|
NM_006761
|
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr12_-_53994783
|
0.007
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr13_+_29598746
|
0.006
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr22_+_24951617
|
0.006
|
NM_004175
|
SNRPD3
|
small nuclear ribonucleoprotein D3 polypeptide 18kDa
|
|
chr22_+_36044412
|
0.005
|
NM_030641
|
APOL6
|
apolipoprotein L, 6
|
|
chr17_-_40190043
|
0.005
|
NM_001242704
|
ZNF385C
|
zinc finger protein 385C
|
|
chr20_+_43104513
|
0.004
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr9_-_38069207
|
0.004
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr12_-_21548370
|
0.004
|
NM_134431
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr12_+_72057676
|
0.004
|
NM_031435
|
THAP2
|
THAP domain containing, apoptosis associated protein 2
|
|
chr13_+_53191601
|
0.004
|
NM_001011724
NM_001011725
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr4_+_184020343
|
0.003
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr6_+_76458888
|
0.003
|
NM_004999
|
MYO6
|
myosin VI
|
|
chr22_-_33402648
|
0.003
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr18_+_77794345
|
0.003
|
NM_001171967
NM_024805
|
RBFA
|
ribosome binding factor A (putative)
|
|
chr11_-_6640661
|
0.003
|
NM_000391
|
TPP1
|
tripeptidyl peptidase I
|
|
chrX_+_146993450
|
0.002
|
NM_001185075
NM_001185076
NM_001185081
NM_001185082
NM_002024
|
FMR1
|
fragile X mental retardation 1
|
|
chr17_-_8021806
|
0.002
|
NM_021628
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr3_+_193310921
|
0.002
|
NM_015560
NM_130831
NM_130832
NM_130833
NM_130834
NM_130835
NM_130836
NM_130837
|
OPA1
|
optic atrophy 1 (autosomal dominant)
|
|
chr4_-_47840058
|
0.002
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr4_-_113437327
|
0.002
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr3_+_187943192
|
0.002
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_-_206950895
|
0.001
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr8_+_59323822
|
0.001
|
NM_001077619
|
UBXN2B
|
UBX domain protein 2B
|
|
chr11_+_109964086
|
0.001
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr6_+_42788793
|
0.001
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr3_+_143690639
|
0.000
|
NM_173552
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr2_+_75873907
|
0.000
|
NM_014763
|
MRPL19
|
mitochondrial ribosomal protein L19
|