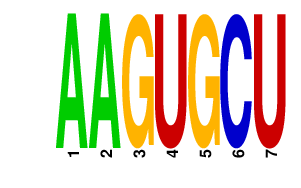
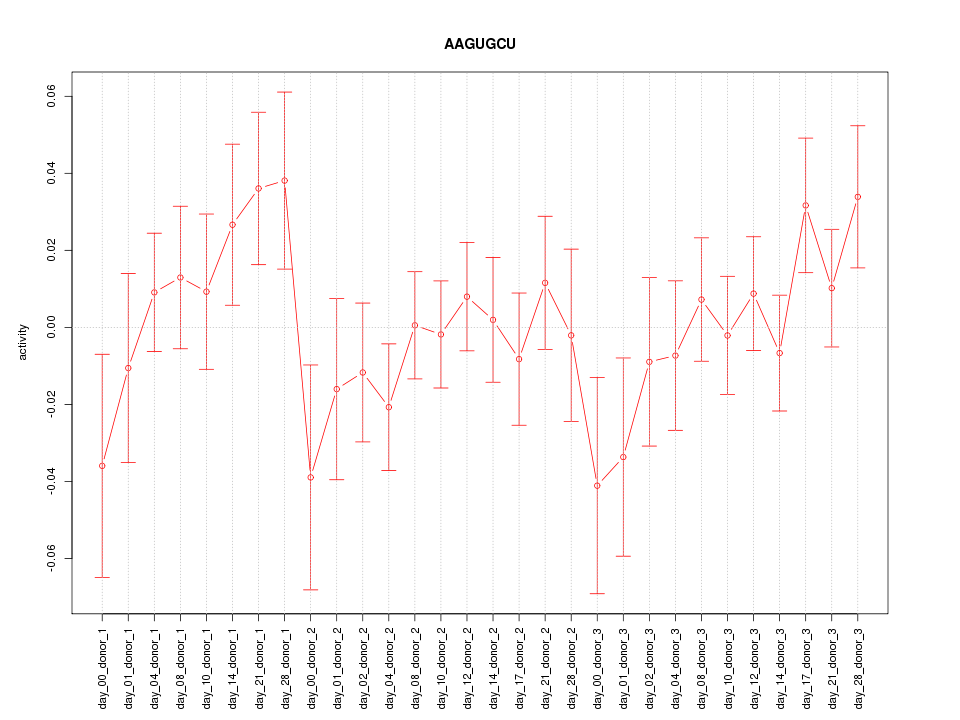
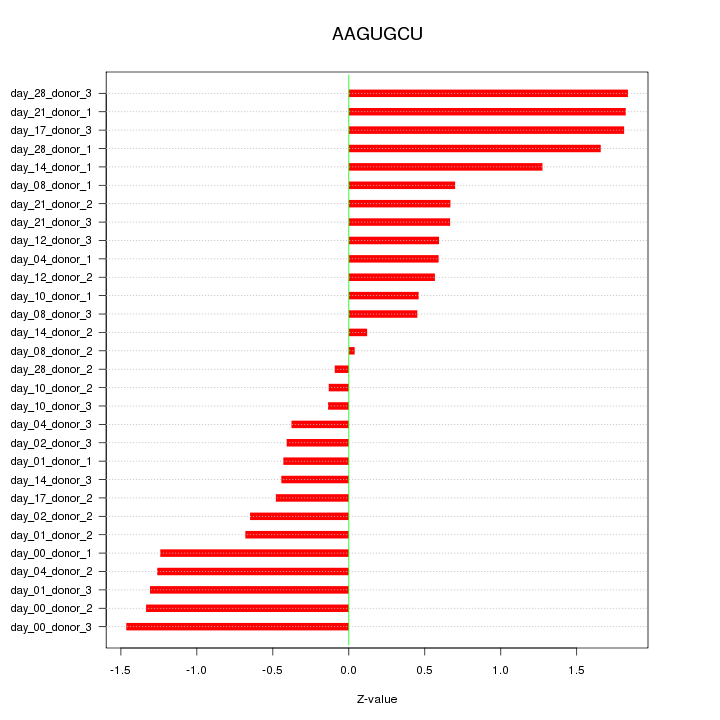
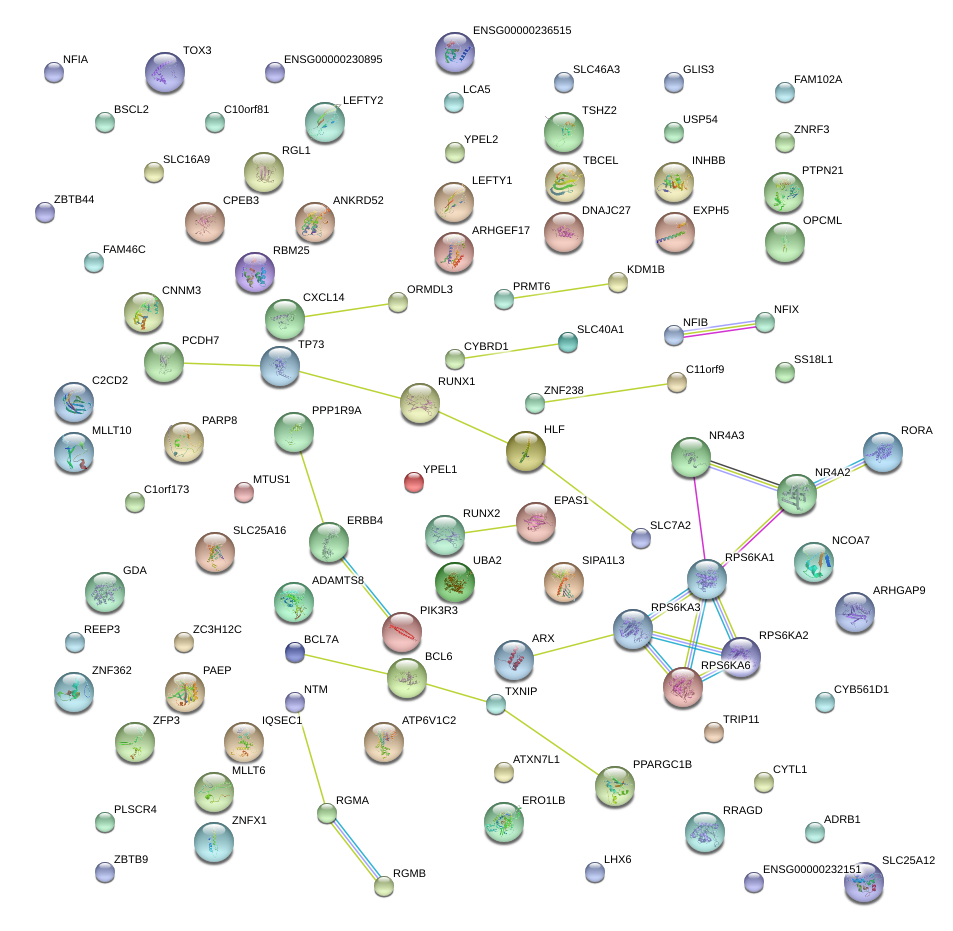
|
chr16_-_52580805
|
5.620
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr16_-_52581713
|
5.390
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr1_-_75139421
|
3.771
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr1_+_183605207
|
3.449
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr9_-_4300028
|
3.217
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr6_-_90121966
|
3.140
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr9_-_4152182
|
3.112
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr3_-_145968958
|
2.792
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr2_+_121103670
|
2.729
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr10_+_115803805
|
2.616
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr9_-_124984018
|
2.551
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr9_-_124990949
|
2.425
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr1_+_61547625
|
2.403
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr5_-_134914968
|
2.378
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr9_-_124990706
|
2.308
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr1_+_61542928
|
2.255
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr15_-_93632161
|
2.087
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93616891
|
2.044
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr9_+_74729510
|
2.038
|
NM_001242507
|
GDA
|
guanine deaminase
|
|
chr15_-_93617091
|
1.925
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr1_+_244214552
|
1.915
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_61547388
|
1.884
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr15_-_93616358
|
1.849
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93631751
|
1.806
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr1_+_3607232
|
1.746
|
NM_001126240
NM_001126241
NM_001126242
NM_001204189
NM_001204190
NM_001204191
|
TP73
|
tumor protein p73
|
|
chr1_+_3569098
|
1.726
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr20_+_51588945
|
1.706
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr2_-_190445521
|
1.706
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr1_+_244216506
|
1.697
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_118148506
|
1.670
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr9_-_124989755
|
1.667
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr1_+_3614609
|
1.618
|
NM_001204192
|
TP73
|
tumor protein p73
|
|
chr3_-_13008959
|
1.616
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr2_+_97481977
|
1.572
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr20_+_51801821
|
1.572
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr22_+_29279573
|
1.560
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr17_+_4981721
|
1.544
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr21_-_43373938
|
1.522
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr5_+_149109814
|
1.509
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr6_+_33422319
|
1.473
|
NM_152735
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
|
chr5_+_149151502
|
1.463
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr11_-_108464344
|
1.440
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr6_+_18155535
|
1.400
|
NM_153042
|
KDM1B
|
lysine (K)-specific demethylase 1B
|
|
chr13_-_29292961
|
1.389
|
NM_001135919
NM_181785
|
SLC46A3
|
solute carrier family 46, member 3
|
|
chr2_+_172378856
|
1.384
|
NM_001127383
NM_024843
|
CYBRD1
|
cytochrome b reductase 1
|
|
chr14_-_89020888
|
1.351
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr9_+_74764266
|
1.339
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr2_-_157189040
|
1.328
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr1_-_226128872
|
1.313
|
NM_001172425
NM_003240
|
LEFTY2
|
left-right determination factor 2
|
|
chr6_-_167040700
|
1.271
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr1_-_46598250
|
1.268
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr10_+_115511212
|
1.267
|
NM_001193434
NM_001193435
NM_024889
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr6_-_167275657
|
1.264
|
NM_001006932
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr15_-_61521478
|
1.237
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_-_130742803
|
1.198
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr10_-_61469322
|
1.198
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr7_+_94536926
|
1.155
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr12_-_56652118
|
1.142
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr1_-_236445294
|
1.130
|
NM_019891
|
ERO1LB
|
ERO1-like beta (S. cerevisiae)
|
|
chr14_+_73525187
|
1.121
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr17_+_53342320
|
1.107
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr15_-_60884615
|
1.100
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr20_+_60718473
|
1.093
|
NM_198935
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr2_-_213403280
|
1.063
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr1_+_107599266
|
1.026
|
NM_018137
|
PRMT6
|
protein arginine methyltransferase 6
|
|
chr7_+_94539209
|
1.017
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr5_+_98104998
|
1.016
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr6_+_126111782
|
1.012
|
NM_001122842
NM_001199621
NM_181782
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr8_-_17555158
|
1.009
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_-_80247103
|
0.999
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr17_+_36861857
|
0.997
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr11_-_130184459
|
0.981
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr15_-_77712436
|
0.969
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr15_-_60919642
|
0.967
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr8_-_17580024
|
0.964
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_+_49962771
|
0.959
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr7_-_105319608
|
0.957
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr3_-_187454247
|
0.957
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr14_-_92506284
|
0.952
|
NM_004239
|
TRIP11
|
thyroid hormone receptor interactor 11
|
|
chr2_+_10861740
|
0.937
|
NM_001039362
NM_144583
|
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2
|
|
chr3_-_187452669
|
0.936
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr22_+_29313376
|
0.932
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr11_-_132812870
|
0.927
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr11_+_120894756
|
0.921
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chr17_+_57408907
|
0.919
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr3_-_13114547
|
0.919
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr2_-_25194772
|
0.909
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr11_+_61520000
|
0.900
|
NM_001127392
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr8_-_17579729
|
0.884
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr8_+_17354562
|
0.876
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr8_-_17658385
|
0.865
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr21_-_36421586
|
0.852
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chrX_-_25034064
|
0.852
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr6_+_126240369
|
0.833
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr11_+_73019662
|
0.832
|
NM_014786
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr12_+_122459791
|
0.818
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chrX_-_20284708
|
0.818
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr11_+_109964086
|
0.815
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr1_+_145438437
|
0.811
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr11_-_62475060
|
0.808
|
NM_001122955
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr1_+_33722158
|
0.790
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr9_-_130712866
|
0.787
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr20_-_47894591
|
0.787
|
NM_021035
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr1_+_110036657
|
0.784
|
NM_001134402
NM_001134400
NM_001134403
NM_001134404
NM_182580
|
CYB561D1
|
cytochrome b-561 domain containing 1
|
|
chr10_-_94050799
|
0.782
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr4_+_30721865
|
0.775
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr10_+_65281108
|
0.774
|
NM_001001330
|
REEP3
|
receptor accessory protein 3
|
|
chr19_+_38397841
|
0.772
|
NM_015073
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3
|
|
chr2_-_172750724
|
0.764
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr17_-_38083762
|
0.760
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr5_+_74807580
|
0.757
|
NM_016218
|
POLK
|
polymerase (DNA directed) kappa
|
|
chr4_-_170924911
|
0.751
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr17_-_47439326
|
0.749
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr3_-_46037299
|
0.747
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr8_+_28351694
|
0.746
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr3_-_187463474
|
0.746
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr12_+_121647659
|
0.743
|
NM_002560
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4
|
|
chr4_-_53525316
|
0.740
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr10_+_92980322
|
0.735
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr14_+_58765209
|
0.723
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr3_-_15106806
|
0.723
|
NM_022497
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr1_-_235490801
|
0.720
|
NM_001206794
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr9_-_37465384
|
0.712
|
NM_014872
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr3_+_23987514
|
0.711
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr1_+_26438267
|
0.709
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr3_+_23986735
|
0.701
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr8_+_17396285
|
0.701
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr4_+_86851425
|
0.697
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr11_+_61522860
|
0.696
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr4_-_125633716
|
0.696
|
NM_001167882
NM_020337
|
ANKRD50
|
ankyrin repeat domain 50
|
|
chr5_+_140186658
|
0.684
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr5_+_140254930
|
0.680
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr20_+_30865435
|
0.678
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr16_-_71748697
|
0.678
|
NM_015020
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr5_+_140213968
|
0.671
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr17_-_40897025
|
0.669
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|
|
chr1_+_2985730
|
0.666
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr2_-_174828775
|
0.665
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr5_+_140220811
|
0.662
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr11_+_35684342
|
0.656
|
NM_017583
|
TRIM44
|
tripartite motif containing 44
|
|
chr11_+_118307072
|
0.653
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr10_-_94003002
|
0.652
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr7_-_2354050
|
0.650
|
NM_013321
|
SNX8
|
sorting nexin 8
|
|
chr10_+_102295572
|
0.647
|
NM_017902
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr16_-_2185824
|
0.644
|
NM_000296
NM_001009944
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr4_+_86396251
|
0.640
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr3_-_113160318
|
0.639
|
NM_001164496
NM_018338
|
WDR52
|
WD repeat domain 52
|
|
chr2_-_128100673
|
0.637
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr4_+_6271558
|
0.624
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr21_-_43346780
|
0.621
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr6_+_126102278
|
0.612
|
NM_001199619
NM_001199620
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr16_-_3493489
|
0.606
|
NM_152457
|
ZNF597
|
zinc finger protein 597
|
|
chr5_+_133706869
|
0.606
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B
|
|
chr6_-_143266283
|
0.604
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr17_-_27621163
|
0.586
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr19_-_45004482
|
0.586
|
NM_013256
|
ZNF180
|
zinc finger protein 180
|
|
chr15_+_96869156
|
0.585
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chrX_-_24045302
|
0.581
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr2_-_225450105
|
0.581
|
NM_003590
|
CUL3
|
cullin 3
|
|
chr5_+_49961732
|
0.580
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chrX_+_119384490
|
0.580
|
NM_006777
NM_001184742
|
ZBTB33
|
zinc finger and BTB domain containing 33
|
|
chr5_+_56110899
|
0.577
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr15_+_31619043
|
0.573
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr7_-_6746541
|
0.570
|
NM_006956
NM_016265
|
ZNF12
|
zinc finger protein 12
|
|
chr2_-_48132656
|
0.569
|
NM_001190274
|
FBXO11
|
F-box protein 11
|
|
chr18_-_19284737
|
0.567
|
NM_138340
|
ABHD3
|
abhydrolase domain containing 3
|
|
chr5_+_102201526
|
0.566
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr1_-_235491494
|
0.565
|
NM_016374
NM_031371
|
ARID4B
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr14_-_53620032
|
0.560
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr4_+_57775064
|
0.553
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr5_+_131705353
|
0.549
|
NM_003060
|
SLC22A5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5
|
|
chr5_+_140247767
|
0.542
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr13_+_78271820
|
0.538
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chrX_-_99986496
|
0.534
|
NM_001174068
NM_080737
|
SYTL4
|
synaptotagmin-like 4
|
|
chr15_+_96876568
|
0.528
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr6_-_154677868
|
0.526
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr11_-_77531750
|
0.517
|
NM_016578
|
RSF1
|
remodeling and spacing factor 1
|
|
chr8_-_23540401
|
0.517
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr9_+_82186846
|
0.516
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr7_-_152133071
|
0.515
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr5_-_133968463
|
0.514
|
NM_001033503
NM_016103
|
SAR1B
|
SAR1 homolog B (S. cerevisiae)
|
|
chr1_-_36851484
|
0.514
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr11_+_33279167
|
0.502
|
NM_001048200
NM_005734
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr6_-_154651214
|
0.497
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr9_-_115652961
|
0.494
|
NM_033051
|
SLC46A2
|
solute carrier family 46, member 2
|
|
chr4_-_16228097
|
0.492
|
NM_153365
|
TAPT1
|
transmembrane anterior posterior transformation 1
|
|
chr2_-_158731622
|
0.490
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr12_-_89918531
|
0.490
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr20_+_33292117
|
0.490
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr1_+_162467594
|
0.483
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chrX_-_99987072
|
0.476
|
NM_001129896
|
SYTL4
|
synaptotagmin-like 4
|
|
chr1_-_207224298
|
0.476
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chr9_-_127952037
|
0.474
|
NM_001123355
NM_001123369
NM_002721
|
PPP6C
|
protein phosphatase 6, catalytic subunit
|
|
chr4_-_37687932
|
0.473
|
NM_001085399
NM_001085400
|
RELL1
|
RELT-like 1
|
|
chr17_+_73780842
|
0.470
|
NM_001080419
|
UNK
|
unkempt homolog (Drosophila)
|
|
chr4_-_170947428
|
0.466
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr11_-_62473502
|
0.466
|
NM_001130702
NM_032667
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr17_+_7608413
|
0.466
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr17_+_7184978
|
0.466
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr2_-_240322620
|
0.458
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|