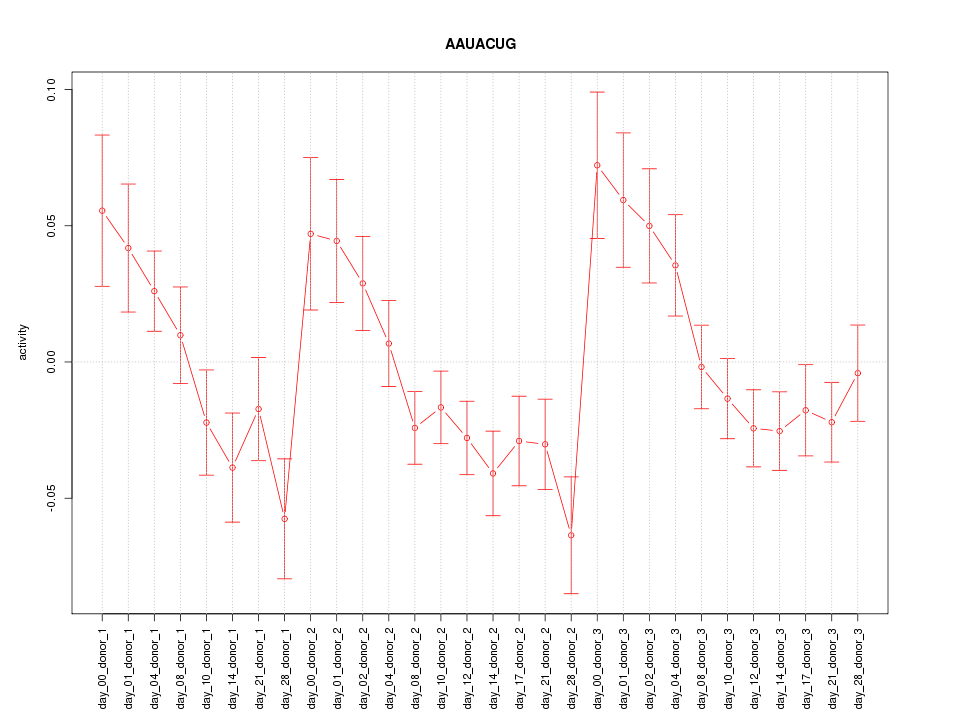
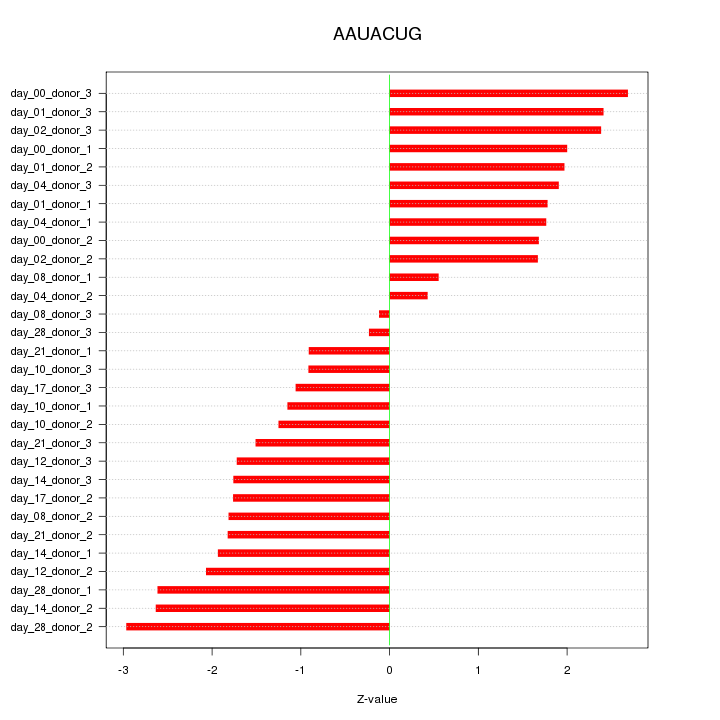
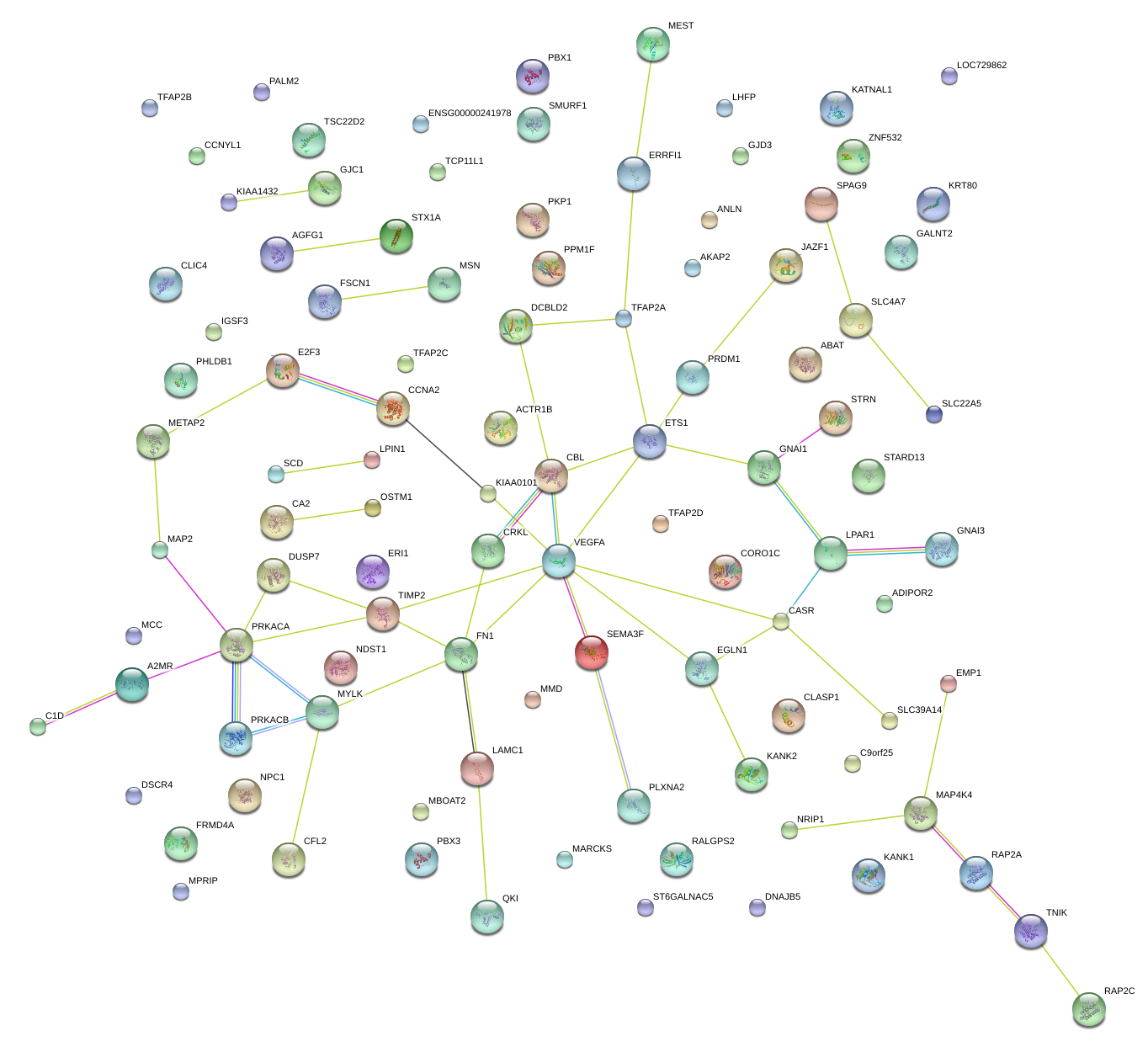
|
chr6_-_30655671
|
6.811
|
NM_133471
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr6_-_30655078
|
6.259
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr10_+_102106771
|
6.095
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr7_+_5632435
|
5.781
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr18_-_21166440
|
5.234
|
NM_000271
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr2_+_210444364
|
5.164
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr2_+_210288668
|
4.864
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chrX_+_64887510
|
4.839
|
NM_002444
|
MSN
|
moesin
|
|
chr11_-_128457452
|
4.631
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr15_-_64673569
|
4.414
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr1_-_8086392
|
4.325
|
NM_018948
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr11_-_128392061
|
4.291
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr9_+_34990266
|
4.250
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr2_+_11886739
|
4.236
|
NM_145693
|
LPIN1
|
lipin 1
|
|
chr18_+_56530060
|
4.120
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr7_+_36429388
|
3.854
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr2_+_102314162
|
3.665
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr9_+_34989637
|
3.637
|
NM_001135004
NM_012266
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr1_+_182992329
|
3.473
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr12_-_52585686
|
3.383
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr9_+_112887780
|
3.355
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr17_+_16945766
|
3.353
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr1_+_178694273
|
3.283
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr2_+_208576239
|
3.217
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr11_+_119076974
|
3.166
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr16_+_8806825
|
3.145
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr21_-_16437125
|
3.121
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr7_-_73133917
|
3.081
|
NM_001165903
NM_004603
|
STX1A
|
syntaxin 1A (brain)
|
|
chr3_-_52090090
|
3.059
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr7_-_28220342
|
3.038
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr12_+_1800188
|
2.985
|
NM_024551
|
ADIPOR2
|
adiponectin receptor 2
|
|
chr5_-_112824526
|
2.983
|
NM_001085377
|
MCC
|
mutated in colorectal cancers
|
|
chr9_+_112810877
|
2.977
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr10_-_14372807
|
2.913
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr3_+_150126705
|
2.828
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr1_+_230202928
|
2.815
|
NM_004481
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2)
|
|
chr16_+_8814567
|
2.797
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr1_+_84630575
|
2.771
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr13_-_30881556
|
2.761
|
NM_032116
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr2_-_216300770
|
2.728
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr5_-_112630557
|
2.684
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr2_-_9143742
|
2.665
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr3_-_27498244
|
2.655
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr17_-_76921453
|
2.609
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_+_163835668
|
2.548
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr9_+_706744
|
2.524
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr1_+_201252579
|
2.499
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr1_-_208417635
|
2.453
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr17_-_42907606
|
2.444
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr22_+_21271619
|
2.444
|
NM_005207
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr1_+_110091185
|
2.442
|
NM_006496
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr9_-_34458477
|
2.435
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr9_+_112542496
|
2.433
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr6_+_106534188
|
2.407
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr1_-_117210301
|
2.401
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr9_+_128510477
|
2.396
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr9_+_128509616
|
2.381
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr9_-_113800243
|
2.339
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr13_-_30881141
|
2.337
|
NM_001014380
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr3_+_50192847
|
2.325
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr3_-_123339423
|
2.309
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr6_+_20402040
|
2.299
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr1_-_231560789
|
2.273
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr12_+_57522247
|
2.248
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr6_-_108395939
|
2.235
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr1_+_84609941
|
2.232
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr6_+_43737937
|
2.229
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_+_25071759
|
2.216
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr1_+_84630014
|
2.204
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_+_130131921
|
2.169
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr6_-_10415438
|
2.146
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr13_-_40177355
|
2.119
|
NM_005780
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr8_+_86376130
|
2.097
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr12_-_109125289
|
2.093
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr14_-_35183722
|
2.084
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr9_+_504661
|
2.060
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr1_+_84647342
|
2.037
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr14_-_35182944
|
2.037
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr5_+_149887673
|
2.016
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr22_-_22307212
|
2.005
|
NM_014634
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F
|
|
chr1_+_77333158
|
1.978
|
NM_030965
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr2_-_122406994
|
1.965
|
NM_001142273
NM_001142274
NM_001207051
NM_015282
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr7_+_130126035
|
1.953
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr17_-_53499055
|
1.944
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr17_-_42908178
|
1.931
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr8_+_22224997
|
1.926
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr11_+_118477212
|
1.917
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr17_-_49198084
|
1.906
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr19_-_11308184
|
1.905
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chrX_-_131351917
|
1.898
|
NM_021183
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr12_+_13349601
|
1.878
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_-_175161832
|
1.865
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr19_-_11305186
|
1.853
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr8_+_8860313
|
1.852
|
NM_153332
|
ERI1
|
exoribonuclease 1
|
|
chr3_-_98620452
|
1.847
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr3_+_121902529
|
1.829
|
NM_000388
|
CASR
|
calcium-sensing receptor
|
|
chr6_-_10419786
|
1.812
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr7_+_130131172
|
1.807
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_-_98741681
|
1.805
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr11_+_33060962
|
1.795
|
NM_018393
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr2_-_37193609
|
1.782
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr6_+_106546736
|
1.781
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr2_+_228336877
|
1.769
|
NM_001135187
NM_001135188
NM_001135189
NM_004504
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr11_+_33061560
|
1.762
|
NM_001145541
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr16_+_8768403
|
1.752
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr13_-_33859835
|
1.745
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr22_-_36424397
|
1.733
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr4_-_122744978
|
1.731
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr11_+_118478305
|
1.709
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr6_+_114178516
|
1.674
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr9_+_5629110
|
1.670
|
NM_001135920
NM_001206557
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr8_+_32579350
|
1.660
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr12_-_106641675
|
1.637
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr22_-_36236421
|
1.624
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr10_+_96162185
|
1.618
|
NM_015188
|
TBC1D12
|
TBC1 domain family, member 12
|
|
chr12_+_120105656
|
1.617
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr8_-_82024277
|
1.605
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr3_-_123603144
|
1.594
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr9_+_131644780
|
1.573
|
NM_001127244
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr8_+_70405027
|
1.571
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr6_-_10412606
|
1.568
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr10_-_120514669
|
1.563
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr17_-_79885544
|
1.552
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr12_+_56324945
|
1.517
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr19_-_40791291
|
1.512
|
NM_001243027
NM_001243028
NM_001626
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr10_-_23003443
|
1.511
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr15_+_40509628
|
1.511
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr2_+_201170603
|
1.510
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr2_-_47143250
|
1.504
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr17_-_71640226
|
1.498
|
NM_001144952
|
SDK2
|
sidekick cell adhesion molecule 2
|
|
chr7_+_4721722
|
1.493
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chrX_-_54522318
|
1.490
|
NM_004463
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr3_+_44379923
|
1.483
|
NM_001029839
NM_173826
|
C3orf23
|
chromosome 3 open reading frame 23
|
|
chr19_+_34287662
|
1.477
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr9_+_131644368
|
1.474
|
NM_001127245
NM_019594
|
LRRC8A
|
leucine rich repeat containing 8 family, member A
|
|
chr14_-_39572414
|
1.473
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr13_-_74707913
|
1.471
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr1_-_120190389
|
1.471
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr11_+_128563772
|
1.466
|
NM_001167681
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr5_+_38845962
|
1.465
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr4_-_153303663
|
1.456
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr7_+_77167351
|
1.439
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr3_-_185826748
|
1.415
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr4_-_139163222
|
1.403
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr8_+_90769974
|
1.392
|
NM_003821
|
RIPK2
|
receptor-interacting serine-threonine kinase 2
|
|
chr13_-_33760215
|
1.391
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr9_+_132815984
|
1.381
|
NM_001136557
NM_001136558
NM_020960
|
GPR107
|
G protein-coupled receptor 107
|
|
chr10_-_105614952
|
1.376
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr9_+_112403067
|
1.373
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr9_-_139440237
|
1.368
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr13_-_33780142
|
1.367
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_-_120169493
|
1.366
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr1_-_214724974
|
1.365
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr7_-_42276617
|
1.364
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr22_-_50913270
|
1.352
|
NM_002972
|
SBF1
|
SET binding factor 1
|
|
chr7_+_28338939
|
1.341
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_+_26205490
|
1.339
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr10_-_99052429
|
1.338
|
NM_001204300
NM_032900
|
ARHGAP19
|
Rho GTPase activating protein 19
|
|
chr4_-_176923572
|
1.332
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr10_-_101945686
|
1.331
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr13_-_21635703
|
1.321
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr2_-_47142950
|
1.312
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr2_+_198669316
|
1.294
|
NM_006226
|
PLCL1
|
phospholipase C-like 1
|
|
chr3_+_158991035
|
1.291
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_+_79439432
|
1.277
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr12_-_51591949
|
1.273
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr10_-_101945787
|
1.269
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr5_+_70751435
|
1.252
|
NM_018429
|
BDP1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB
|
|
chrX_+_150151757
|
1.243
|
NM_005342
|
HMGB3
|
high mobility group box 3
|
|
chr3_-_15140652
|
1.237
|
NM_022340
|
ZFYVE20
|
zinc finger, FYVE domain containing 20
|
|
chr16_-_67281398
|
1.236
|
NM_013241
|
FHOD1
|
formin homology 2 domain containing 1
|
|
chr7_+_138916230
|
1.229
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr2_+_201170795
|
1.227
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr12_+_26126687
|
1.222
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr11_-_46939908
|
1.222
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr8_+_106331146
|
1.220
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr14_-_99737821
|
1.219
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr2_+_23608297
|
1.218
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr14_+_55518313
|
1.210
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr7_+_77166750
|
1.208
|
NM_002835
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr7_+_28725597
|
1.197
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr21_-_15755439
|
1.192
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr9_-_23826062
|
1.191
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr19_-_18632886
|
1.181
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr11_-_35441099
|
1.180
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr3_-_156272909
|
1.173
|
NM_007107
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma)
|
|
chr15_+_72766665
|
1.170
|
NM_005744
|
ARIH1
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila)
|
|
chr5_-_171433876
|
1.162
|
NM_012300
NM_033644
NM_033645
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr17_-_56084656
|
1.159
|
NM_001078166
NM_006924
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr5_+_7396030
|
1.156
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr8_+_32405727
|
1.155
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr13_-_107187312
|
1.121
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr2_-_47168902
|
1.119
|
NM_001171508
NM_001171511
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr21_+_40177847
|
1.113
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr4_-_73434467
|
1.100
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chrX_-_15873094
|
1.099
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr8_+_11561710
|
1.095
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr4_-_68566847
|
1.089
|
NM_018227
|
UBA6
|
ubiquitin-like modifier activating enzyme 6
|
|
chr1_-_225616556
|
1.081
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr9_+_33817168
|
1.073
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|