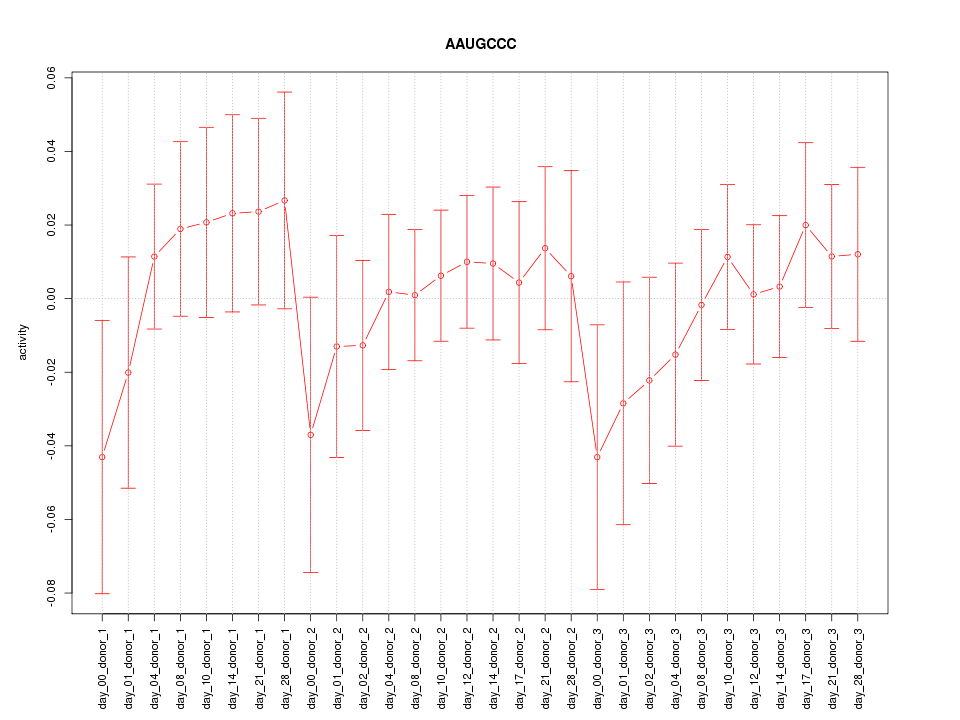
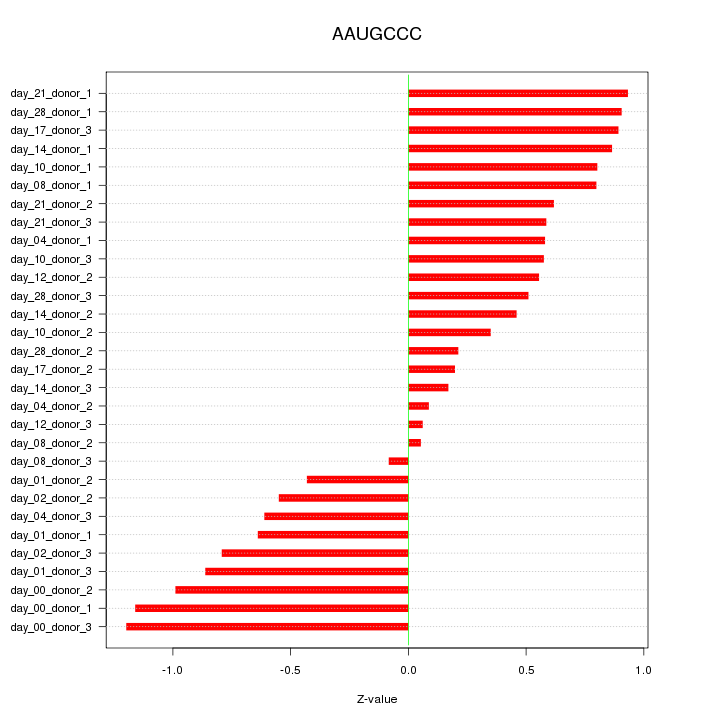
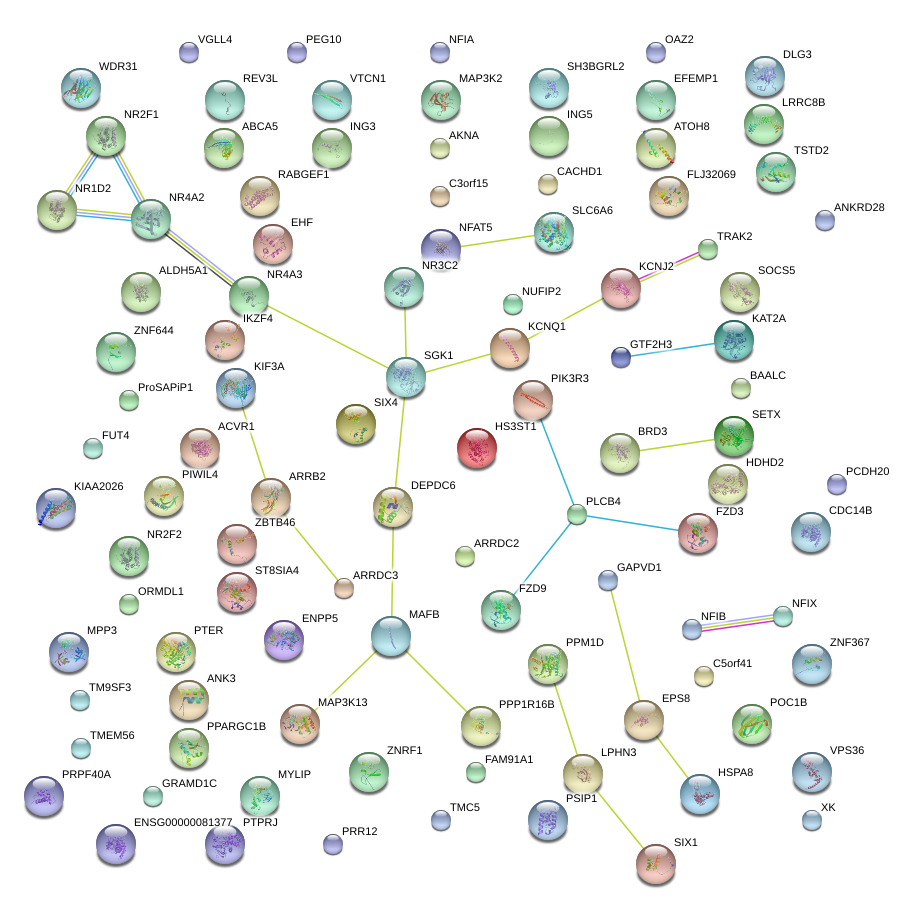
|
chr4_-_149363498
|
2.191
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr20_+_9049660
|
1.578
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr20_+_9288446
|
1.498
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr20_+_9198036
|
1.405
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr16_+_19467771
|
1.402
|
NM_024780
|
TMC5
|
transmembrane channel-like 5
|
|
chr1_+_61547625
|
1.343
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr20_-_39317875
|
1.288
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr1_+_61542928
|
1.287
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr6_-_134496006
|
1.202
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134497069
|
1.182
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr9_-_117156684
|
1.132
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr5_+_149109814
|
1.120
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr16_+_19422056
|
1.111
|
NM_001105248
NM_001105249
|
TMC5
|
transmembrane channel-like 5
|
|
chr5_+_149151502
|
1.103
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_+_61547388
|
1.069
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr12_-_89918531
|
1.058
|
NM_003774
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4)
|
|
chr6_-_134639195
|
1.042
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134498973
|
1.038
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr20_+_37434294
|
0.821
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr3_+_23986735
|
0.820
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_+_23987514
|
0.803
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_+_113557679
|
0.801
|
NM_017577
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr2_+_85980600
|
0.792
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr1_-_46598250
|
0.783
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr6_-_46138584
|
0.765
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr3_+_113616317
|
0.763
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr1_-_117753476
|
0.762
|
NM_001253850
NM_024626
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr17_+_68165660
|
0.756
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr2_-_158731622
|
0.701
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr6_+_16129285
|
0.691
|
NM_013262
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr11_+_2466171
|
0.686
|
NM_000218
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1
|
|
chr17_+_4613780
|
0.636
|
NM_004313
NM_199004
|
ARRB2
|
arrestin, beta 2
|
|
chr12_+_74931550
|
0.622
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr15_-_64995459
|
0.578
|
NM_002537
|
OAZ2
|
ornithine decarboxylase antizyme 2
|
|
chr2_-_158732341
|
0.568
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr2_-_157189040
|
0.557
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr20_-_3149070
|
0.540
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr1_+_64936475
|
0.532
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr9_-_116102533
|
0.527
|
NM_001012361
NM_145241
|
WDR31
|
WD repeat domain 31
|
|
chr5_-_90679115
|
0.513
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr6_+_24495166
|
0.512
|
NM_001080
NM_170740
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1
|
|
chr17_-_27621163
|
0.468
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr12_-_89920038
|
0.466
|
NM_001199781
NM_001199782
NM_172240
|
POC1B-GALNT4
POC1B
|
POC1B-GALNT4 readthrough
POC1 centriolar protein homolog B (Chlamydomonas)
|
|
chr3_-_15900928
|
0.443
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr8_+_28351694
|
0.399
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr10_-_98346793
|
0.398
|
NM_020123
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr6_-_111804413
|
0.391
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr16_+_69599868
|
0.387
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr6_+_80340971
|
0.387
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr13_-_61989654
|
0.386
|
NM_022843
|
PCDH20
|
protocadherin 20
|
|
chr2_-_153573858
|
0.381
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr9_-_99329092
|
0.380
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr5_-_100238870
|
0.379
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr19_+_18118921
|
0.378
|
NM_015683
|
ARRDC2
|
arrestin domain containing 2
|
|
chr9_-_100395961
|
0.377
|
NM_139246
|
TSTD2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2
|
|
chr3_+_14444083
|
0.364
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr11_+_34642587
|
0.360
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr12_-_15942350
|
0.358
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr10_-_61900659
|
0.355
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr10_+_16478941
|
0.352
|
NM_001001484
NM_030664
|
PTER
|
phosphotriesterase related
|
|
chr14_-_61190458
|
0.351
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr8_+_120885899
|
0.350
|
NM_022783
|
DEPTOR
|
DEP domain containing MTOR-interacting protein
|
|
chr10_-_62149487
|
0.349
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr3_-_15839674
|
0.343
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_+_48002063
|
0.343
|
NM_001098503
NM_002843
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr19_+_13105970
|
0.343
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_+_18111923
|
0.339
|
NM_001025604
|
ARRDC2
|
arrestin domain containing 2
|
|
chr3_+_185000714
|
0.336
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr20_-_62436855
|
0.334
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr1_-_91487769
|
0.330
|
NM_032186
NM_201269
|
ZNF644
|
zinc finger protein 644
|
|
chr2_+_242641310
|
0.329
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr10_-_62332666
|
0.323
|
NM_001204404
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr12_+_56414688
|
0.320
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr3_+_119421865
|
0.320
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr10_-_62493282
|
0.312
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_50094911
|
0.299
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chrX_+_69674914
|
0.293
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr9_-_99382073
|
0.289
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr9_-_135230358
|
0.285
|
NM_015046
|
SETX
|
senataxin
|
|
chr9_-_136933091
|
0.285
|
NM_007371
|
BRD3
|
bromodomain containing 3
|
|
chr17_-_40273304
|
0.284
|
NM_021078
|
KAT2A
|
K(lysine) acetyltransferase 2A
|
|
chr15_-_77712436
|
0.283
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr18_-_44676837
|
0.277
|
NM_032124
|
HDHD2
|
haloacid dehalogenase-like hydrolase domain containing 2
|
|
chr9_-_6007891
|
0.270
|
NM_001017969
|
KIAA2026
|
KIAA2026
|
|
chrX_+_69664704
|
0.269
|
NM_021120
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr7_+_94285620
|
0.267
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr5_+_172483285
|
0.267
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr3_-_11761946
|
0.264
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr2_-_190649075
|
0.261
|
NM_001128150
NM_016467
|
ORMDL1
|
ORM1-like 1 (S. cerevisiae)
|
|
chr17_+_58677489
|
0.260
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr7_+_94285676
|
0.260
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr4_-_11430502
|
0.257
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr1_-_91487013
|
0.256
|
NM_016620
|
ZNF644
|
zinc finger protein 644
|
|
chr13_-_53024730
|
0.256
|
NM_016075
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae)
|
|
chr7_+_120590816
|
0.255
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr17_-_67323185
|
0.250
|
NM_172232
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr11_+_94277005
|
0.250
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr8_+_104152876
|
0.249
|
NM_001024372
NM_024812
|
BAALC
|
brain and acute leukemia, cytoplasmic
|
|
chr9_+_128024109
|
0.246
|
NM_015635
|
GAPVD1
|
GTPase activating protein and VPS9 domains 1
|
|
chr5_-_132073143
|
0.243
|
NM_007054
|
KIF3A
|
kinesin family member 3A
|
|
chr3_-_11610264
|
0.243
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_95558072
|
0.241
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr3_+_185080835
|
0.240
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr11_+_34654010
|
0.239
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chrX_+_37545008
|
0.237
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr8_+_124780742
|
0.234
|
NM_144963
|
FAM91A1
|
family with sequence similarity 91, member A1
|
|
chr11_-_122932844
|
0.228
|
NM_006597
NM_153201
|
HSPA8
|
heat shock 70kDa protein 8
|
|
chr9_-_15510209
|
0.224
|
NM_021144
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr3_-_11685395
|
0.224
|
NM_001128219
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_89990396
|
0.221
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr14_-_61115906
|
0.219
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr2_+_46926049
|
0.219
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr16_+_75032874
|
0.218
|
NM_032268
|
ZNRF1
|
zinc and ring finger 1
|
|
chr3_-_11623829
|
0.217
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr7_+_66093854
|
0.210
|
NM_001167961
NM_153033
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr2_-_128100673
|
0.210
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr2_-_202316227
|
0.209
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr2_-_56151273
|
0.209
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr12_+_124118312
|
0.206
|
NM_001516
|
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa
|
|
chr17_-_67311773
|
0.206
|
NM_018672
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr1_+_95582827
|
0.201
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr15_+_96869156
|
0.200
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr16_-_66730466
|
0.197
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chrX_+_69672110
|
0.195
|
NM_020730
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr8_-_103876390
|
0.191
|
NM_015878
NM_148174
|
AZIN1
|
antizyme inhibitor 1
|
|
chr6_+_30295007
|
0.190
|
NM_172016
|
TRIM39
|
tripartite motif containing 39
|
|
chr15_-_89089792
|
0.188
|
NM_001144074
NM_017996
|
DET1
|
de-etiolated homolog 1 (Arabidopsis)
|
|
chr5_+_112312392
|
0.182
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr1_-_78225319
|
0.181
|
NM_015017
NM_201624
NM_201626
|
USP33
|
ubiquitin specific peptidase 33
|
|
chr15_+_96876568
|
0.181
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr4_+_88928797
|
0.178
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr13_-_21476895
|
0.172
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr7_+_22766765
|
0.169
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr8_+_67687415
|
0.169
|
NM_013257
NM_170709
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr2_+_46524448
|
0.168
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr3_-_33481862
|
0.168
|
NM_001128160
NM_014517
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr12_-_7281374
|
0.167
|
NM_031491
|
RBP5
|
retinol binding protein 5, cellular
|
|
chr1_+_90024271
|
0.166
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr9_-_14314036
|
0.165
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr2_+_46926323
|
0.165
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr12_+_25348138
|
0.165
|
NM_001001660
|
LYRM5
|
LYR motif containing 5
|
|
chr20_+_33292117
|
0.164
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr7_-_150675236
|
0.162
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr8_+_67624652
|
0.162
|
NM_001033578
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3
|
|
chr6_+_30294620
|
0.161
|
NM_021253
|
TRIM39
|
tripartite motif containing 39
|
|
chr12_+_121124922
|
0.160
|
NM_014730
|
MLEC
|
malectin
|
|
chr3_-_33481877
|
0.157
|
NM_001128161
|
UBP1
|
upstream binding protein 1 (LBP-1a)
|
|
chr22_-_41842853
|
0.157
|
NM_016272
|
TOB2
|
transducer of ERBB2, 2
|
|
chr17_+_61627818
|
0.156
|
NM_005828
|
DCAF7
|
DDB1 and CUL4 associated factor 7
|
|
chr2_-_240322620
|
0.155
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr19_+_19431622
|
0.153
|
NM_015329
|
MAU2
|
MAU2 chromatid cohesion factor homolog (C. elegans)
|
|
chr1_+_101361589
|
0.152
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr1_+_145096391
|
0.151
|
NM_004892
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chrX_-_54209639
|
0.150
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chr2_+_131862419
|
0.146
|
NM_001100623
NM_017958
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2
|
|
chr6_+_43138919
|
0.146
|
NM_003131
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr3_-_112218383
|
0.146
|
NM_001085357
NM_181780
|
BTLA
|
B and T lymphocyte associated
|
|
chr1_-_39338976
|
0.146
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr1_+_2985730
|
0.145
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr17_-_16118789
|
0.145
|
NM_006311
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr14_+_52734430
|
0.144
|
NM_000953
|
PTGDR
|
prostaglandin D2 receptor (DP)
|
|
chr17_-_27278471
|
0.143
|
NM_001033561
NM_020889
|
PHF12
|
PHD finger protein 12
|
|
chr22_+_40440664
|
0.143
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_-_15798057
|
0.141
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_-_27742224
|
0.140
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_+_64073040
|
0.138
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr13_-_41240607
|
0.135
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr10_+_181410
|
0.134
|
NM_001202464
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr12_-_49449106
|
0.133
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr16_+_14013989
|
0.133
|
NM_005236
|
ERCC4
|
excision repair cross-complementing rodent repair deficiency, complementation group 4
|
|
chr2_-_200322699
|
0.130
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr20_-_62284701
|
0.130
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr20_+_60697471
|
0.128
|
NM_144703
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae)
|
|
chr6_-_139613114
|
0.127
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr11_-_27741192
|
0.124
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr15_+_96875793
|
0.124
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_+_2140449
|
0.123
|
NM_175607
|
CNTN4
|
contactin 4
|
|
chr11_-_27721975
|
0.119
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr10_-_32217762
|
0.119
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr3_-_47823386
|
0.118
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr18_-_45456925
|
0.116
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr14_+_55493837
|
0.116
|
NM_080867
NM_199421
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr9_-_127905715
|
0.116
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr10_+_119000583
|
0.115
|
NM_003054
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr1_+_203274646
|
0.115
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr11_-_27723061
|
0.115
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_-_57021891
|
0.114
|
NM_001010862
|
SPIN3
|
spindlin family, member 3
|
|
chr6_-_109804432
|
0.113
|
NM_001164313
NM_014797
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr1_-_154600446
|
0.112
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr3_+_45730677
|
0.112
|
NM_014016
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr7_+_101460881
|
0.111
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr16_-_57219951
|
0.109
|
NM_024946
|
FAM192A
|
family with sequence similarity 192, member A
|
|
chr7_-_141401889
|
0.108
|
NM_001080392
|
KIAA1147
|
KIAA1147
|
|
chr17_-_44302716
|
0.108
|
NM_001193465
|
KIAA1267
|
KIAA1267
|
|
chr12_+_68042493
|
0.107
|
NM_003583
NM_006482
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr11_-_68609358
|
0.106
|
NM_001031847
NM_001876
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver)
|
|
chr18_-_45457454
|
0.106
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr1_+_236958539
|
0.106
|
NM_000254
|
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase
|
|
chr5_-_53606387
|
0.105
|
NM_019087
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr12_-_46384365
|
0.104
|
NM_004719
|
SCAF11
|
SR-related CTD-associated factor 11
|