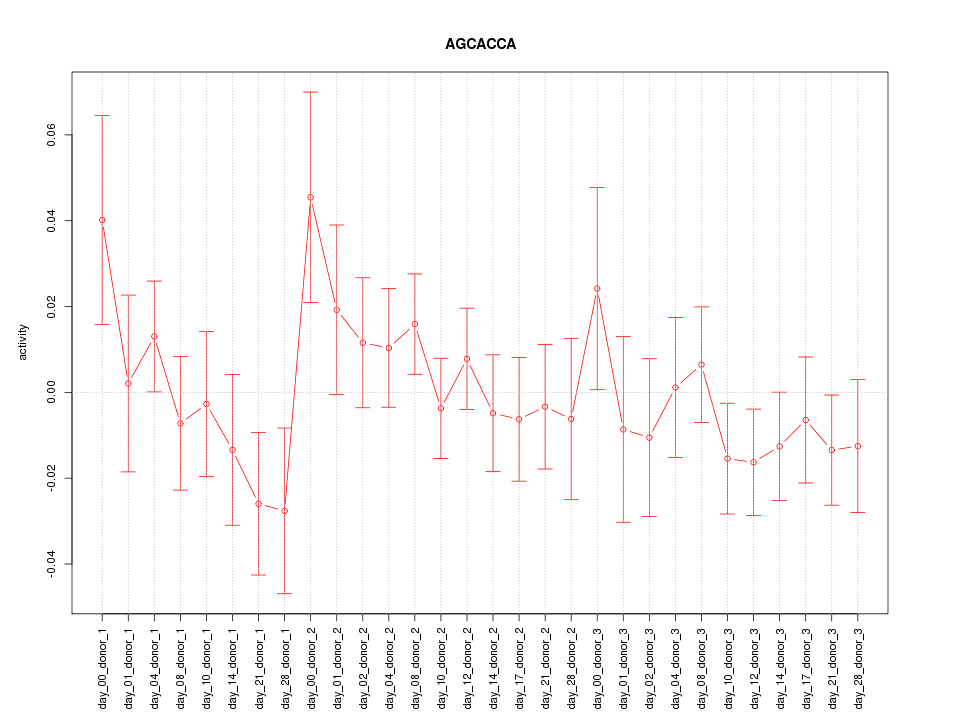
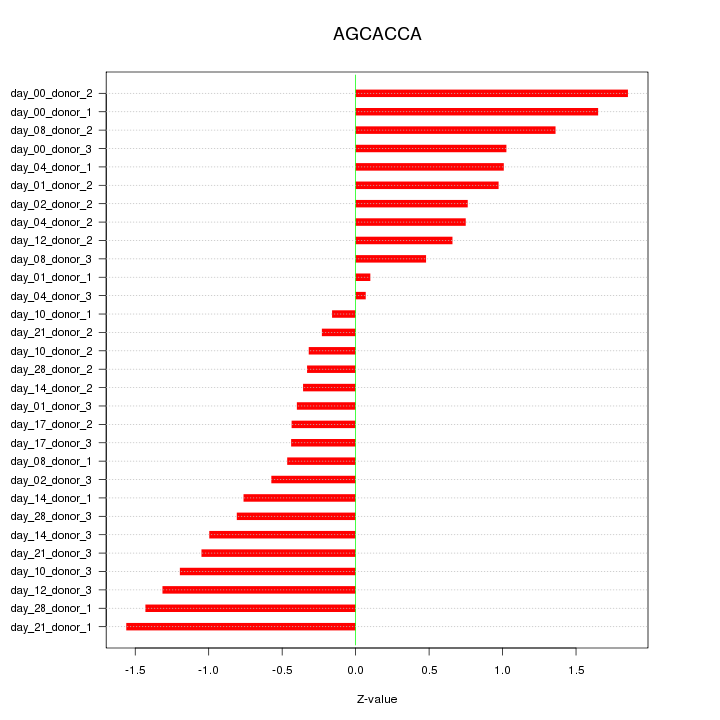
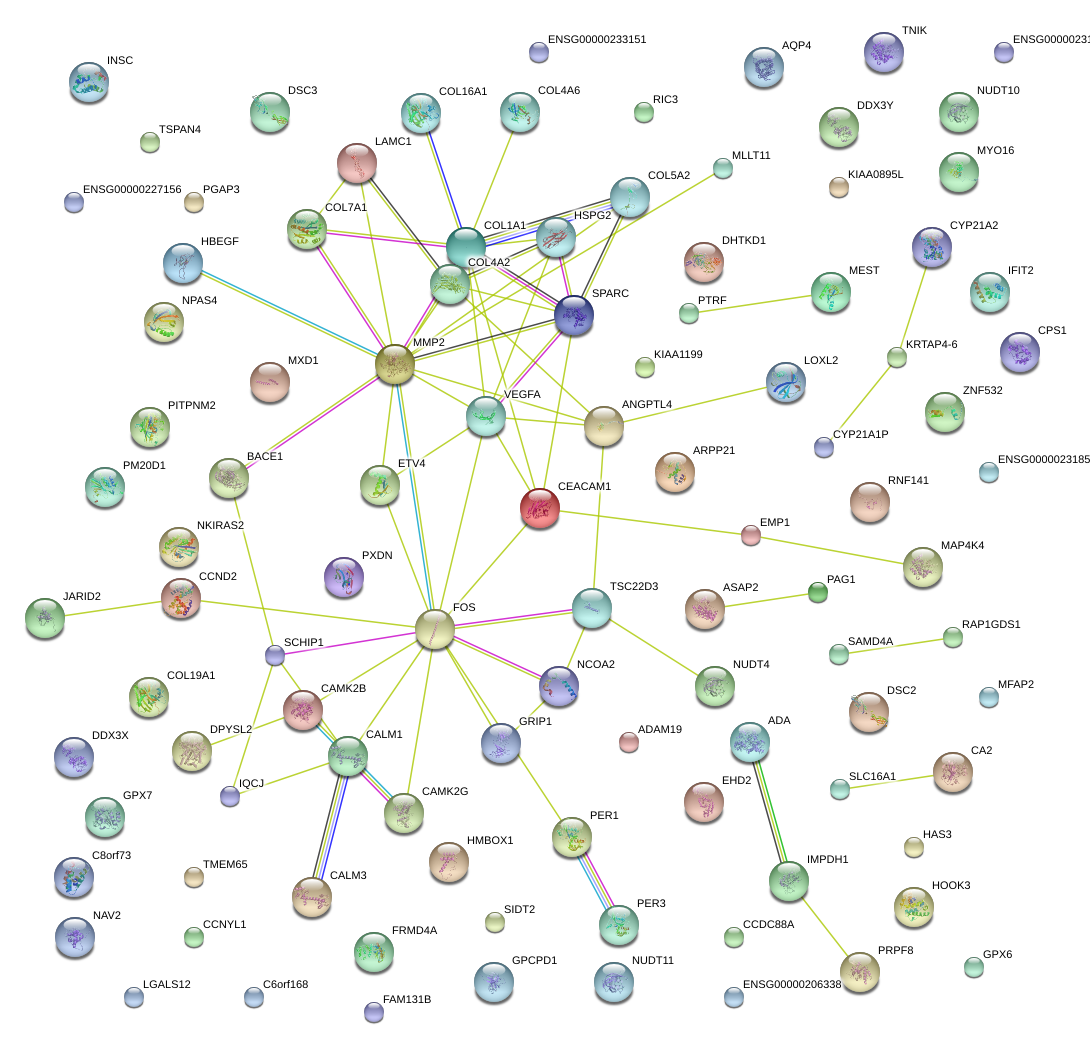
|
chr8_-_23261675
|
2.917
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr2_-_1748285
|
2.663
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr13_+_110959592
|
2.522
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr5_-_151066492
|
2.515
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr1_-_17307107
|
2.502
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr1_-_17308072
|
2.343
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr1_+_151032150
|
2.263
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr16_+_69139466
|
2.204
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr1_-_113498828
|
2.194
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr15_+_81071708
|
2.178
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr16_+_55515468
|
2.163
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_-_113498615
|
2.108
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr16_+_69141442
|
2.097
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr8_+_86376130
|
2.088
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr12_+_13349601
|
2.067
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr2_-_190044330
|
2.051
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_-_48632592
|
2.015
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr5_-_139726173
|
1.991
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chrX_-_107681555
|
1.924
|
NM_033641
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chrX_-_107682600
|
1.821
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr16_+_55512801
|
1.709
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr18_+_56530060
|
1.604
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr19_+_48216600
|
1.556
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr14_+_75745480
|
1.498
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr7_+_130131921
|
1.498
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_130126035
|
1.430
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_-_143059696
|
1.426
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr20_-_43280359
|
1.409
|
NM_000022
|
ADA
|
adenosine deaminase
|
|
chr8_-_82024277
|
1.375
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chrX_-_106959597
|
1.370
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chrX_-_51239426
|
1.360
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr7_-_143059144
|
1.339
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr18_-_28681885
|
1.308
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr17_-_48278987
|
1.296
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr7_+_130131172
|
1.288
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr14_+_55034329
|
1.271
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr6_-_99797530
|
1.261
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr12_-_67072881
|
1.259
|
NM_001178074
NM_021150
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr2_-_55646946
|
1.248
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr1_-_22263676
|
1.238
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr13_+_109281569
|
1.234
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr12_+_4382882
|
1.229
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr8_+_26371461
|
1.223
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr17_-_40575117
|
1.221
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr1_+_182992329
|
1.207
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr7_-_128050006
|
1.193
|
NM_000883
NM_001102605
NM_001142576
NM_183243
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr5_-_157002739
|
1.189
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr20_-_5591487
|
1.184
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr10_-_75634219
|
1.168
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr11_-_8190535
|
1.167
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr17_-_8055684
|
1.141
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr13_+_109248499
|
1.139
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr2_+_208576239
|
1.130
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr19_-_43032525
|
1.123
|
NM_001024912
NM_001184813
NM_001184815
NM_001184816
NM_001205344
NM_001712
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr17_-_41623652
|
1.122
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr3_+_158991035
|
1.102
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr8_+_26435339
|
1.085
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr6_+_43737937
|
1.072
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr2_+_102314162
|
1.071
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr18_-_24442534
|
1.070
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr1_-_32169619
|
1.064
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr2_+_211342405
|
1.064
|
NM_001122633
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr17_-_41623246
|
1.048
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr4_+_99182520
|
1.036
|
NM_001100426
NM_001100427
NM_001100428
NM_001100429
NM_001100430
NM_021159
|
RAP1GDS1
|
RAP1, GTP-GDP dissociation stimulator 1
|
|
chr11_+_844445
|
1.029
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr10_-_14372807
|
1.028
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr19_+_8429010
|
1.020
|
NM_001039667
NM_139314
|
ANGPTL4
|
angiopoietin-like 4
|
|
chr11_+_842821
|
1.006
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr8_+_42752019
|
1.001
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr2_+_211458078
|
1.000
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr7_-_128045995
|
1.000
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr1_+_7844713
|
0.998
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr11_-_117186918
|
0.977
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr12_-_123594974
|
0.967
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr2_+_211421322
|
0.955
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr11_-_117166247
|
0.945
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr17_+_40169592
|
0.945
|
NM_001001349
|
NKIRAS2
|
NFKB inhibitor interacting Ras-like 2
|
|
chr2_+_9346876
|
0.945
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chrX_-_106960268
|
0.934
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_+_19372270
|
0.933
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr2_+_70142152
|
0.931
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr19_+_47104490
|
0.926
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr8_-_144654921
|
0.924
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr10_+_12110921
|
0.915
|
NM_018706
|
DHTKD1
|
dehydrogenase E1 and transketolase domain containing 1
|
|
chr11_-_10562689
|
0.913
|
NM_016422
|
RNF141
|
ring finger protein 141
|
|
chr14_+_55221505
|
0.907
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr11_+_844062
|
0.903
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr11_+_117049838
|
0.882
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr6_+_15246299
|
0.879
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr18_-_24445652
|
0.878
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chrX_-_107019001
|
0.877
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr6_+_70576447
|
0.868
|
NM_001858
|
COL19A1
|
collagen, type XIX, alpha 1
|
|
chr16_-_67217825
|
0.864
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr8_-_125384916
|
0.857
|
NM_194291
|
TMEM65
|
transmembrane protein 65
|
|
chr1_+_53068042
|
0.856
|
NM_015696
|
GPX6
GPX7
|
glutathione peroxidase 6 (olfactory)
glutathione peroxidase 7
|
|
chr3_+_159557649
|
0.853
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_+_66188474
|
0.846
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chrY_+_15016018
|
0.822
|
NM_001122665
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr7_-_73133917
|
0.804
|
NM_001165903
NM_004603
|
STX1A
|
syntaxin 1A (brain)
|
|
chr17_+_40172086
|
0.800
|
NM_001144927
NM_001144928
NM_001144929
NM_017595
|
NKIRAS2
|
NFKB inhibitor interacting Ras-like 2
|
|
chr7_-_28220342
|
0.798
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr3_+_182970853
|
0.796
|
NM_032047
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr17_+_26662547
|
0.791
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr1_+_40627031
|
0.790
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr22_-_37584254
|
0.787
|
NM_031910
NM_182486
|
C1QTNF6
|
C1q and tumor necrosis factor related protein 6
|
|
chr14_-_45431121
|
0.782
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chrX_-_109683457
|
0.777
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr22_-_39095991
|
0.773
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr11_+_19734821
|
0.770
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr4_-_52904424
|
0.767
|
NM_000232
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein)
|
|
chr22_-_39636906
|
0.765
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr2_-_238322840
|
0.755
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr18_+_29027731
|
0.749
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr19_+_34287662
|
0.744
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr13_-_110959441
|
0.742
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr12_+_32654976
|
0.739
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr20_+_42295708
|
0.739
|
NM_002466
|
MYBL2
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 2
|
|
chr16_+_90015827
|
0.736
|
NM_001242819
|
DEF8
|
differentially expressed in FDCP 8 homolog (mouse)
|
|
chr16_-_19729417
|
0.733
|
NM_001012991
|
C16orf88
|
chromosome 16 open reading frame 88
|
|
chr4_+_124317884
|
0.732
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr10_-_92617670
|
0.731
|
NM_000872
NM_019859
NM_019860
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr19_+_34855594
|
0.729
|
NM_001184722
NM_000175
|
GPI
|
glucose-6-phosphate isomerase
|
|
chr2_-_220094354
|
0.726
|
NM_001077198
NM_024085
|
ATG9A
|
ATG9 autophagy related 9 homolog A (S. cerevisiae)
|
|
chr6_-_43596919
|
0.721
|
NM_019096
|
GTPBP2
|
GTP binding protein 2
|
|
chr15_+_42066631
|
0.710
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr14_+_24838301
|
0.706
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr3_-_57678594
|
0.702
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr3_-_120169493
|
0.698
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr17_+_2699731
|
0.698
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr11_+_60691911
|
0.692
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chrX_-_109561314
|
0.691
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr4_+_113066552
|
0.690
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr6_-_33548017
|
0.669
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr16_-_23160509
|
0.659
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr15_+_79724857
|
0.645
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr11_+_111944967
|
0.640
|
NM_001082969
NM_001082970
NM_018195
|
C11orf57
|
chromosome 11 open reading frame 57
|
|
chr4_+_144257982
|
0.636
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr11_-_118781612
|
0.631
|
NM_182557
|
BCL9L
|
B-cell CLL/lymphoma 9-like
|
|
chr12_-_133464197
|
0.629
|
NM_001161344
NM_001161345
NM_001161346
NM_001161347
NM_018223
|
CHFR
|
checkpoint with forkhead and ring finger domains
|
|
chr15_-_72598655
|
0.627
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr16_-_56459371
|
0.626
|
NM_001144
|
AMFR
|
autocrine motility factor receptor
|
|
chr21_-_32931289
|
0.626
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr6_+_53883713
|
0.621
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr1_-_27816676
|
0.620
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr22_-_39640956
|
0.620
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr16_+_28986024
|
0.614
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr15_-_76005179
|
0.609
|
NM_001897
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr4_-_165304406
|
0.604
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chrX_+_129139163
|
0.603
|
NM_021946
|
BCORL1
|
BCL6 corepressor-like 1
|
|
chr2_-_230933618
|
0.603
|
NM_152527
|
SLC16A14
|
solute carrier family 16, member 14 (monocarboxylic acid transporter 14)
|
|
chr10_+_101419169
|
0.601
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr20_-_25604583
|
0.601
|
NM_152667
|
NANP
|
N-acetylneuraminic acid phosphatase
|
|
chr2_-_37193609
|
0.593
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr19_-_16738986
|
0.588
|
NM_004831
|
MED26
|
mediator complex subunit 26
|
|
chr15_+_68570038
|
0.587
|
NM_015322
|
FEM1B
|
fem-1 homolog b (C. elegans)
|
|
chr5_+_149887673
|
0.583
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr2_+_173292306
|
0.579
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chrX_-_64254592
|
0.578
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr13_+_28194846
|
0.577
|
NM_001206559
|
POLR1D
|
polymerase (RNA) I polypeptide D, 16kDa
|
|
chr9_+_5629110
|
0.574
|
NM_001135920
NM_001206557
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr10_-_33224485
|
0.566
|
NM_033668
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr10_+_89623091
|
0.564
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr17_-_7298160
|
0.564
|
NM_001201576
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr4_-_164534656
|
0.564
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chrX_-_71351750
|
0.554
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr10_-_29923900
|
0.552
|
NM_021738
|
SVIL
|
supervillin
|
|
chrX_+_107683015
|
0.551
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr5_-_141704575
|
0.551
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr6_-_30043547
|
0.549
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr1_+_203595897
|
0.547
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr13_-_74707913
|
0.547
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr12_+_123868608
|
0.540
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr10_+_105036783
|
0.540
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr14_+_94492683
|
0.537
|
NM_023112
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr8_+_22022644
|
0.532
|
NM_001199
NM_006129
|
BMP1
|
bone morphogenetic protein 1
|
|
chr5_+_14664764
|
0.531
|
NM_138348
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr8_-_93029864
|
0.530
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_74056042
|
0.527
|
NM_201647
|
STAMBP
|
STAM binding protein
|
|
chr1_-_229478687
|
0.524
|
NM_145257
|
C1orf96
|
chromosome 1 open reading frame 96
|
|
chr14_+_24836144
|
0.522
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr4_-_153303663
|
0.522
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr5_+_66124603
|
0.521
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr7_-_25019625
|
0.519
|
NM_015550
NM_145320
NM_145321
NM_145322
|
OSBPL3
|
oxysterol binding protein-like 3
|
|
chr6_-_80656870
|
0.516
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr9_-_14910992
|
0.515
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr9_+_112887780
|
0.515
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr12_+_50898744
|
0.513
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr1_-_150849211
|
0.508
|
NM_001197325
NM_001668
NM_178427
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr2_-_31637598
|
0.505
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr1_-_68299047
|
0.504
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr2_+_74056095
|
0.504
|
NM_006463
NM_213622
|
STAMBP
|
STAM binding protein
|
|
chr2_-_162931051
|
0.503
|
NM_001935
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr4_+_124320664
|
0.501
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr8_-_93111915
|
0.499
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_134326030
|
0.498
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr5_-_174871162
|
0.495
|
NM_000794
|
DRD1
|
dopamine receptor D1
|
|
chr8_+_1772095
|
0.493
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr8_+_38585703
|
0.492
|
NM_001146216
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr7_+_5632435
|
0.489
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr1_-_217804312
|
0.488
|
NM_018040
|
GPATCH2
|
G patch domain containing 2
|