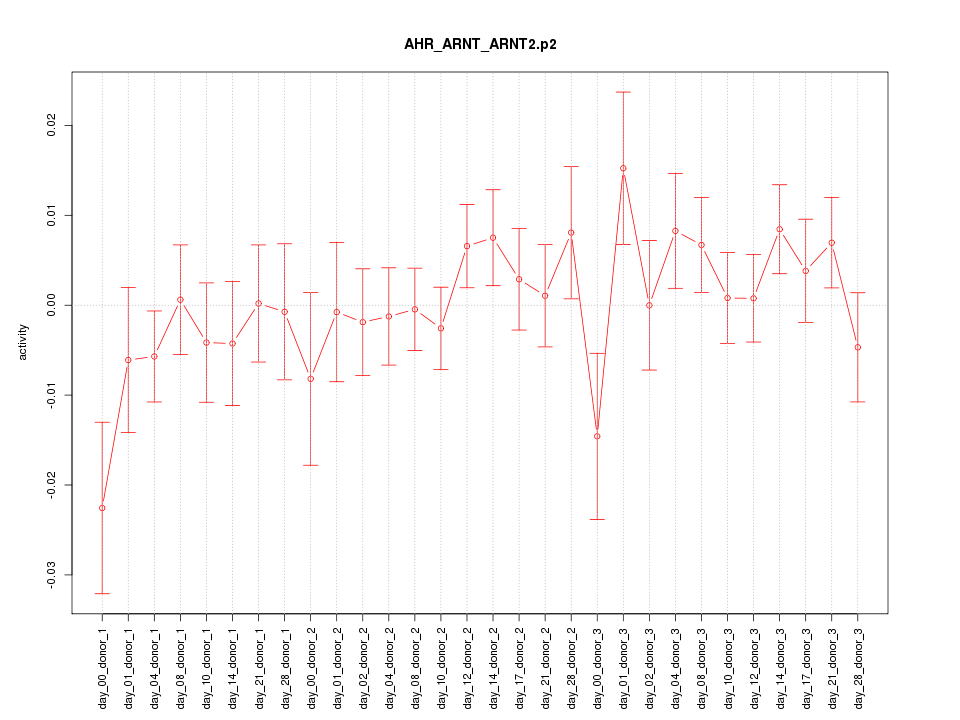
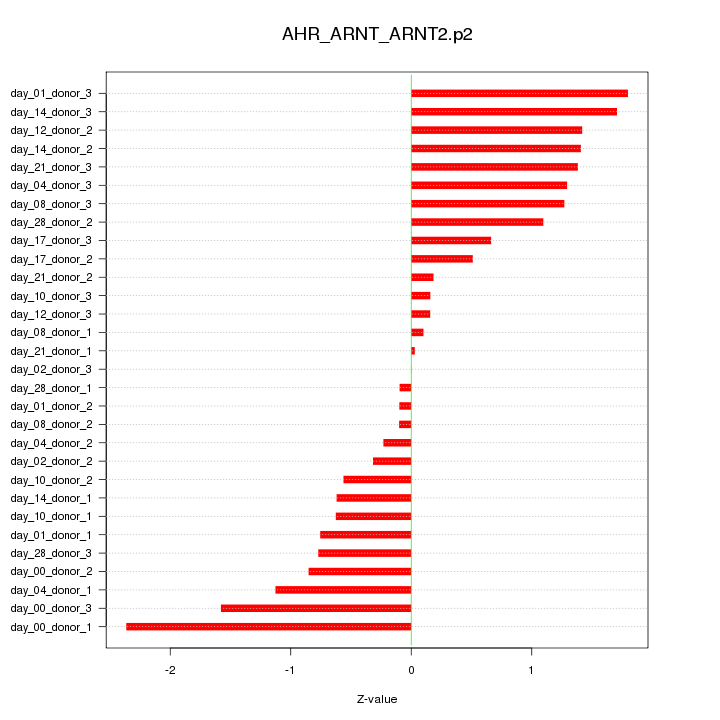
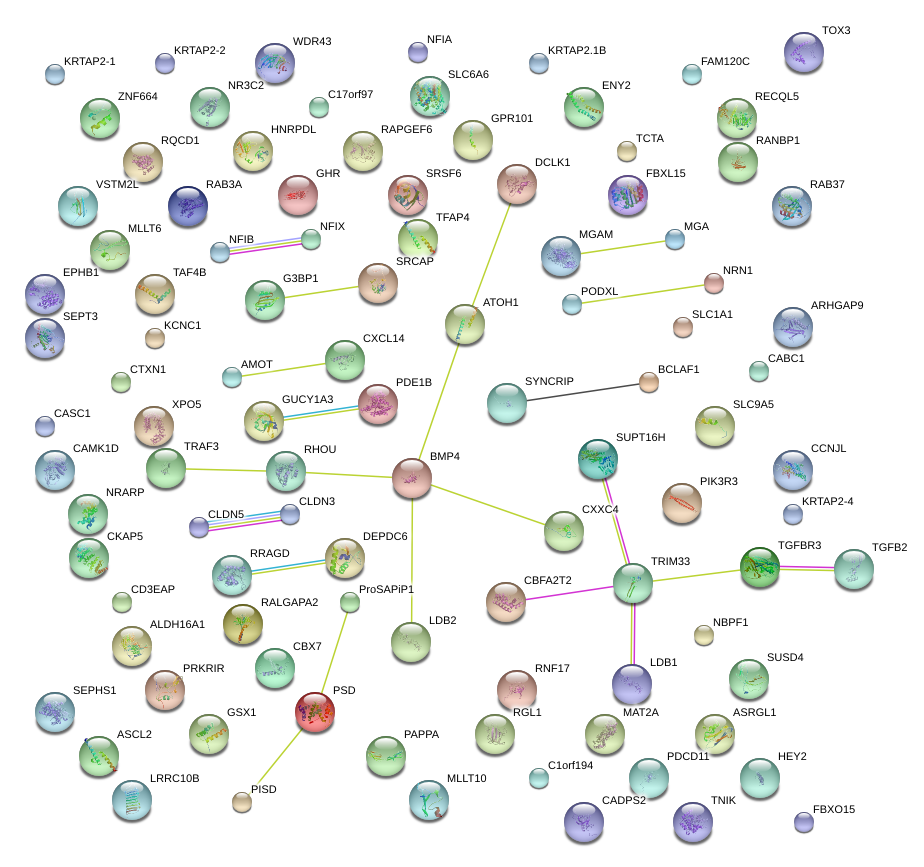
|
chr20_+_42086533
|
2.593
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr3_+_49449753
|
2.518
|
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr3_+_49449638
|
2.245
|
NM_022171
|
TCTA
|
T-cell leukemia translocation altered gene
|
|
chr1_+_61547625
|
1.594
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr4_-_149363498
|
1.581
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr11_-_46867788
|
1.414
|
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr22_+_42372763
|
1.400
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr5_-_134914447
|
1.354
|
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr22_-_39548448
|
1.327
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr13_-_36705432
|
1.324
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr11_+_61276271
|
1.279
|
NM_001145077
|
LRRC10B
|
leucine rich repeat containing 10B
|
|
chr10_-_104178569
|
1.274
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr5_+_42423811
|
1.268
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr3_-_45267068
|
1.223
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr1_-_1535446
|
1.199
|
NM_001242659
|
LOC643988
|
uncharacterized LOC643988
|
|
chr1_+_228871109
|
1.113
|
|
RHOU
|
ras homolog gene family, member U
|
|
chr10_-_104178900
|
1.078
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr20_-_3154169
|
1.071
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr4_+_94750041
|
1.065
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chr11_-_2291809
|
1.056
|
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr18_+_23806385
|
1.030
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr6_-_90121656
|
0.996
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chrX_-_136113832
|
0.978
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr5_+_151151483
|
0.978
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr7_-_73184535
|
0.954
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr4_-_105412466
|
0.954
|
NM_025212
|
CXXC4
|
CXXC finger protein 4
|
|
chr17_+_72667255
|
0.940
|
NM_175738
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr5_-_159739482
|
0.927
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr1_-_92351476
|
0.926
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr11_+_62104773
|
0.912
|
NM_001083926
NM_025080
|
ASRGL1
|
asparaginase like 1
|
|
chr3_+_14444083
|
0.911
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr3_-_171177851
|
0.905
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr19_-_18314731
|
0.895
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr10_+_12391728
|
0.894
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr20_+_32077780
|
0.894
|
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr14_+_103243832
|
0.885
|
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr20_+_42086468
|
0.881
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr11_-_2292181
|
0.877
|
NM_005170
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr14_-_54421269
|
0.876
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr2_+_85766291
|
0.871
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr7_-_131241321
|
0.866
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr5_+_151151516
|
0.864
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr14_-_21852085
|
0.856
|
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr1_+_183774214
|
0.849
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr6_-_86352656
|
0.839
|
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr5_-_134914968
|
0.801
|
NM_004887
|
CXCL14
|
chemokine (C-X-C motif) ligand 14
|
|
chr10_-_103879933
|
0.798
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr16_+_30710461
|
0.796
|
NM_006662
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chr17_+_260092
|
0.781
|
NM_001013672
|
C17orf97
|
chromosome 17 open reading frame 97
|
|
chr3_-_45267804
|
0.759
|
NM_015444
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr5_+_151151511
|
0.756
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr5_+_151151514
|
0.753
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr6_+_126070813
|
0.751
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr16_+_30709913
|
0.748
|
|
|
|
|
chr10_+_12391708
|
0.747
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr6_+_126070724
|
0.745
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr1_+_227127959
|
0.744
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr6_-_6007632
|
0.739
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr6_-_136610894
|
0.737
|
|
BCLAF1
|
BCL2-associated transcription factor 1
|
|
chr19_+_45909466
|
0.723
|
NM_012099
|
CD3EAP
|
CD3e molecule, epsilon associated protein
|
|
chr12_-_25348031
|
0.710
|
NM_001082972
NM_001082973
NM_001204101
NM_001204102
NM_018272
|
CASC1
|
cancer susceptibility candidate 1
|
|
chr5_+_151151462
|
0.704
|
NM_005754
NM_198395
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr3_-_45267620
|
0.699
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr22_-_19512001
|
0.699
|
|
CLDN5
|
claudin 5
|
|
chr1_+_218519576
|
0.687
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr9_+_4490193
|
0.687
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr2_+_219433302
|
0.678
|
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr5_+_42424553
|
0.677
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr1_+_2518464
|
0.677
|
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr1_+_2518071
|
0.675
|
NM_001195736
NM_001195737
NM_001195738
NM_001195740
NM_001195741
NM_152371
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr8_+_120885899
|
0.672
|
NM_022783
|
DEPTOR
|
DEP domain containing MTOR-interacting protein
|
|
chr6_-_43543749
|
0.665
|
|
XPO5
|
exportin 5
|
|
chr18_-_71814948
|
0.663
|
NM_152676
NM_001142958
|
FBXO15
|
F-box protein 15
|
|
chr1_-_223537400
|
0.661
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr14_-_21852161
|
0.659
|
|
SUPT16H
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr1_-_46598250
|
0.653
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr16_-_4322695
|
0.651
|
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chrX_-_112084006
|
0.650
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr1_-_109656478
|
0.646
|
NM_001122961
|
C1orf194
|
chromosome 1 open reading frame 194
|
|
chr6_-_86352650
|
0.643
|
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr4_-_83350452
|
0.639
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr6_-_90121966
|
0.638
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr4_+_156588811
|
0.624
|
NM_001130684
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3
|
|
chr5_+_42425093
|
0.615
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr2_+_29117503
|
0.613
|
NM_015131
|
WDR43
|
WD repeat domain 43
|
|
chr15_+_41952572
|
0.609
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr17_-_39203423
|
0.606
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr9_-_140196702
|
0.604
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr20_+_36531548
|
0.602
|
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr11_-_76091866
|
0.599
|
NM_004705
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor)
|
|
chr2_-_73460326
|
0.586
|
NM_032319
|
PRADC1
|
protease-associated domain containing 1
|
|
chrX_-_54209639
|
0.582
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chr16_-_52580805
|
0.579
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr12_+_124457568
|
0.579
|
NM_001204299
NM_001204298
NM_152437
|
ZNF664-FAM101A
ZNF664
|
protein FAM101A
zinc finger protein 664
|
|
chr6_+_126070738
|
0.574
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr20_-_20693122
|
0.573
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr11_+_17757494
|
0.571
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr20_+_36531498
|
0.571
|
NM_080607
|
VSTM2L
|
V-set and transmembrane domain containing 2 like
|
|
chr22_-_19511777
|
0.570
|
|
CLDN5
|
claudin 5
|
|
chr1_+_227127999
|
0.566
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr13_+_28366779
|
0.566
|
NM_145657
|
GSX1
|
GS homeobox 1
|
|
chr7_-_122526781
|
0.564
|
NM_001009571
NM_001167940
NM_017954
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr6_-_86352982
|
0.564
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr10_-_13390189
|
0.563
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr3_+_134514044
|
0.563
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr22_+_20105068
|
0.562
|
|
RANBP1
|
RAN binding protein 1
|
|
chr5_-_130970803
|
0.559
|
|
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr17_+_36861857
|
0.552
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr8_+_110346549
|
0.552
|
NM_001193557
NM_020189
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr1_-_115053677
|
0.551
|
|
TRIM33
|
tripartite motif containing 33
|
|
chr19_-_7990974
|
0.548
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr12_+_54943358
|
0.548
|
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr19_+_49956472
|
0.545
|
NM_001145396
NM_153329
|
ALDH16A1
|
aldehyde dehydrogenase 16 family, member A1
|
|
chr16_+_67282774
|
0.542
|
NM_004594
|
SLC9A5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5
|
|
chr2_-_172016904
|
0.542
|
|
TLK1
|
tousled-like kinase 1
|
|
chr19_-_39340362
|
0.542
|
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr19_-_14316980
|
0.539
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr1_+_1447495
|
0.535
|
NM_001170535
NM_018188
|
ATAD3A
|
ATPase family, AAA domain containing 3A
|
|
chr19_+_49956492
|
0.535
|
|
ALDH16A1
|
aldehyde dehydrogenase 16 family, member A1
|
|
chrX_-_140271275
|
0.534
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr19_-_7293876
|
0.532
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr6_-_43543600
|
0.531
|
|
XPO5
|
exportin 5
|
|
chr8_+_85095452
|
0.531
|
NM_001100392
NM_001100393
|
RALYL
|
RALY RNA binding protein-like
|
|
chr5_+_134181729
|
0.530
|
|
C5orf24
|
chromosome 5 open reading frame 24
|
|
chr17_+_72427535
|
0.529
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr16_+_22825806
|
0.529
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr19_-_19774472
|
0.528
|
|
ATP13A1
|
ATPase type 13A1
|
|
chr19_-_10341947
|
0.528
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr1_+_1447871
|
0.527
|
NM_001170536
|
ATAD3A
|
ATPase family, AAA domain containing 3A
|
|
chr15_-_93632161
|
0.525
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr14_+_60975937
|
0.522
|
NM_007374
|
SIX6
|
SIX homeobox 6
|
|
chr10_+_26505235
|
0.522
|
NM_000818
NM_001134366
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr19_-_19774501
|
0.520
|
NM_020410
|
ATP13A1
|
ATPase type 13A1
|
|
chr8_+_67341244
|
0.519
|
NM_015169
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr6_-_119670897
|
0.519
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr20_+_13976014
|
0.517
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr10_+_105156382
|
0.517
|
NM_014976
|
PDCD11
|
programmed cell death 11
|
|
chr6_-_136610932
|
0.516
|
NM_001077440
NM_001077441
NM_014739
|
BCLAF1
|
BCL2-associated transcription factor 1
|
|
chr6_-_90121911
|
0.516
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr1_+_227127937
|
0.514
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_-_115053758
|
0.514
|
NM_015906
NM_033020
|
TRIM33
|
tripartite motif containing 33
|
|
chr8_+_85095501
|
0.512
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr10_+_105156430
|
0.509
|
|
PDCD11
|
programmed cell death 11
|
|
chr5_+_127419407
|
0.508
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr12_-_9102262
|
0.507
|
NM_001207024
NM_002355
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr15_+_84115979
|
0.506
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr1_+_227127993
|
0.505
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr9_+_33025270
|
0.504
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr16_+_776957
|
0.503
|
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr12_+_110906203
|
0.502
|
NM_013300
|
C12orf24
|
chromosome 12 open reading frame 24
|
|
chr14_+_103243813
|
0.501
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr5_-_81046943
|
0.499
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr4_+_55523906
|
0.499
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr7_+_91763851
|
0.498
|
|
LOC613126
|
uncharacterized LOC613126
|
|
chr2_-_33824294
|
0.495
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr15_-_93631751
|
0.494
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr12_-_88974094
|
0.493
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr20_+_32077904
|
0.491
|
NM_001032999
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr7_+_145813828
|
0.490
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr17_+_53342356
|
0.490
|
|
HLF
|
hepatic leukemia factor
|
|
chr19_-_46088084
|
0.484
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr6_+_43484834
|
0.484
|
|
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa
|
|
chr16_+_77225094
|
0.484
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr13_-_36705513
|
0.481
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr6_+_43484776
|
0.480
|
NM_203290
|
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa
|
|
chr11_+_819563
|
0.476
|
|
PNPLA2
|
patatin-like phospholipase domain containing 2
|
|
chr9_+_33025302
|
0.476
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr12_-_9102193
|
0.472
|
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr2_+_198570027
|
0.468
|
NM_138395
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial
|
|
chr20_+_58179601
|
0.467
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr11_+_73357222
|
0.467
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr5_-_81046858
|
0.467
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr16_+_22825481
|
0.466
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr7_+_44144002
|
0.462
|
|
AEBP1
|
AE binding protein 1
|
|
chr3_-_66024508
|
0.461
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr1_+_205538164
|
0.460
|
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr14_+_77648127
|
0.458
|
|
TMEM63C
|
transmembrane protein 63C
|
|
chr15_+_65822842
|
0.455
|
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1
|
|
chr13_-_95953572
|
0.455
|
NM_001105515
NM_005845
|
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4
|
|
chr1_+_1447537
|
0.454
|
|
ATAD3A
ATAD3B
|
ATPase family, AAA domain containing 3A
ATPase family, AAA domain containing 3B
|
|
chr12_+_110906659
|
0.453
|
|
C12orf24
|
chromosome 12 open reading frame 24
|
|
chr11_+_66624501
|
0.451
|
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr1_+_221052845
|
0.449
|
|
HLX
|
H2.0-like homeobox
|
|
chr12_+_110906532
|
0.448
|
|
C12orf24
|
chromosome 12 open reading frame 24
|
|
chr22_+_45098046
|
0.448
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr11_-_86383355
|
0.447
|
NM_006680
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr2_-_27712520
|
0.447
|
NM_015662
|
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas)
|
|
chr14_-_65569056
|
0.445
|
|
MAX
|
MYC associated factor X
|
|
chr9_-_89562103
|
0.445
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr22_-_29196452
|
0.444
|
|
XBP1
|
X-box binding protein 1
|
|
chr7_-_117513466
|
0.442
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr1_-_92351613
|
0.439
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr16_-_4323000
|
0.439
|
NM_003223
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr13_+_37006408
|
0.438
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr4_-_54232007
|
0.436
|
NM_152540
|
SCFD2
|
sec1 family domain containing 2
|
|
chr1_+_9648931
|
0.434
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr20_-_45318135
|
0.433
|
NM_033550
|
TP53RK
|
TP53 regulating kinase
|
|
chr3_-_72897575
|
0.432
|
|
SHQ1
|
SHQ1 homolog (S. cerevisiae)
|
|
chr6_-_107435635
|
0.428
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr21_-_47743753
|
0.427
|
NM_058180
|
C21orf58
|
chromosome 21 open reading frame 58
|