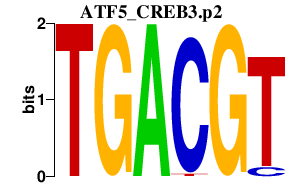
|
chr20_+_58179601
|
4.041
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr8_-_110661007
|
3.246
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr12_+_132629014
|
2.794
|
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr2_-_216300770
|
2.595
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr12_+_132628978
|
2.582
|
NM_024078
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr19_+_50979656
|
2.497
|
NM_175063
NM_206538
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr19_+_50979788
|
2.114
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr5_+_72861567
|
2.041
|
NM_032175
|
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae)
|
|
chr4_-_89618928
|
1.848
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr19_-_9695166
|
1.741
|
NM_001008727
|
ZNF121
|
zinc finger protein 121
|
|
chr9_-_125027017
|
1.730
|
|
RBM18
|
RNA binding motif protein 18
|
|
chr10_+_103911932
|
1.592
|
NM_004741
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr10_+_35484829
|
1.513
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr10_-_97416367
|
1.491
|
|
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chr8_+_40010986
|
1.422
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr17_+_1633754
|
1.402
|
|
WDR81
|
WD repeat domain 81
|
|
chr7_-_25164902
|
1.358
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr11_-_3818736
|
1.318
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr11_+_126138934
|
1.300
|
NM_017547
|
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1
|
|
chr9_-_140082983
|
1.291
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr4_+_75310852
|
1.275
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr20_+_42086533
|
1.263
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr17_+_40985444
|
1.252
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr16_+_19078958
|
1.248
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr12_-_132628841
|
1.232
|
NM_175066
|
DDX51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr2_-_10588451
|
1.231
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr4_+_75480628
|
1.230
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr2_-_10588273
|
1.229
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr19_-_46088021
|
1.228
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr11_+_6502607
|
1.227
|
NM_012192
|
FXC1
|
fracture callus 1 homolog (rat)
|
|
chr10_-_97416564
|
1.197
|
NM_001017423
NM_002860
|
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chr17_+_40985416
|
1.193
|
NM_005789
NM_176863
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr17_+_40985412
|
1.176
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr9_-_125027080
|
1.165
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr3_+_158288952
|
1.149
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chrX_+_24167861
|
1.144
|
|
ZFX
|
zinc finger protein, X-linked
|
|
chr12_+_122150638
|
1.126
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chrX_+_24167728
|
1.119
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr19_-_46088074
|
1.113
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr4_+_56261763
|
1.105
|
|
|
|
|
chr6_-_134496833
|
1.096
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_+_126139022
|
1.092
|
|
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1
|
|
chr11_+_126139049
|
1.090
|
|
FOXRED1
|
FAD-dependent oxidoreductase domain containing 1
|
|
chr10_+_119000583
|
1.079
|
NM_003054
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr4_+_72052354
|
1.073
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr7_+_44646189
|
1.067
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr16_+_19078916
|
1.062
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr7_+_44646197
|
1.057
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr7_+_44646168
|
1.037
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr17_+_40985434
|
1.015
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr5_-_1524022
|
1.007
|
NM_024830
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr16_+_19079220
|
0.976
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr6_-_134497069
|
0.970
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_195808980
|
0.960
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr21_-_34915124
|
0.959
|
NM_001136005
NM_001136006
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr2_-_157189040
|
0.943
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr6_+_64282449
|
0.933
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr17_+_46018888
|
0.922
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr17_+_1619816
|
0.919
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr11_-_111749810
|
0.910
|
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1
|
|
chr4_-_79860369
|
0.909
|
NM_001040202
|
PAQR3
|
progestin and adipoQ receptor family member III
|
|
chr7_+_6144504
|
0.908
|
NM_032172
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr4_-_23891608
|
0.908
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr2_-_60780404
|
0.905
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr11_-_118927815
|
0.902
|
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr20_-_56284945
|
0.895
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr21_-_43430448
|
0.886
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr7_-_25164869
|
0.881
|
|
CYCS
|
cytochrome c, somatic
|
|
chr12_+_66583004
|
0.867
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr19_+_10362886
|
0.867
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr9_+_125027152
|
0.865
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr10_+_99079017
|
0.865
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr13_-_45563554
|
0.864
|
NM_012345
|
NUFIP1
|
nuclear fragile X mental retardation protein interacting protein 1
|
|
chr16_-_68057098
|
0.859
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr18_-_267987
|
0.855
|
NM_005131
|
THOC1
|
THO complex 1
|
|
chr22_-_29196452
|
0.836
|
|
XBP1
|
X-box binding protein 1
|
|
chr17_+_38171613
|
0.832
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr17_+_27071008
|
0.831
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr11_-_3818971
|
0.829
|
NM_005387
NM_016320
NM_139131
NM_139132
|
NUP98
|
nucleoporin 98kDa
|
|
chr1_-_91487543
|
0.828
|
|
ZNF644
|
zinc finger protein 644
|
|
chr12_+_82752275
|
0.825
|
NM_032230
|
C12orf26
|
chromosome 12 open reading frame 26
|
|
chr17_+_40985425
|
0.819
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr8_-_104427319
|
0.815
|
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr20_+_18794369
|
0.813
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr7_-_156685621
|
0.812
|
|
LMBR1
|
limb region 1 homolog (mouse)
|
|
chr17_-_39165343
|
0.808
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr7_+_30174343
|
0.807
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr7_-_8301767
|
0.804
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr12_+_66582914
|
0.795
|
NM_001142523
NM_007199
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr12_+_90103470
|
0.783
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chr4_-_156297944
|
0.780
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr16_-_68057120
|
0.776
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr1_-_53704121
|
0.772
|
NM_002370
|
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila)
|
|
chr6_-_108395939
|
0.771
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr12_+_27397077
|
0.767
|
NM_015000
|
STK38L
|
serine/threonine kinase 38 like
|
|
chr9_+_80850938
|
0.765
|
NM_001098802
NM_032171
|
CEP78
|
centrosomal protein 78kDa
|
|
chr11_-_116968986
|
0.762
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr11_-_77184549
|
0.761
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr17_-_72968750
|
0.758
|
|
|
|
|
chr8_-_10697382
|
0.758
|
|
PINX1
|
PIN2/TERF1 interacting, telomerase inhibitor 1
|
|
chr7_+_116660263
|
0.756
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr7_+_129008044
|
0.754
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_+_113930430
|
0.751
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr1_+_948846
|
0.750
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr7_+_44646220
|
0.748
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr11_-_67276101
|
0.744
|
NM_005851
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr3_-_119182423
|
0.742
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr20_+_35201948
|
0.738
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr12_-_94853453
|
0.733
|
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr19_+_49122762
|
0.728
|
|
SPHK2
|
sphingosine kinase 2
|
|
chr11_+_113930296
|
0.726
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr18_+_7567313
|
0.723
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr3_+_170075449
|
0.723
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr22_-_29196537
|
0.723
|
NM_001079539
NM_005080
|
XBP1
|
X-box binding protein 1
|
|
chr7_+_107531585
|
0.720
|
NM_000108
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr7_+_129007942
|
0.716
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr3_-_195808934
|
0.712
|
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr19_+_49122789
|
0.710
|
NM_001204159
|
SPHK2
|
sphingosine kinase 2
|
|
chr12_+_56660913
|
0.702
|
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr19_-_46088084
|
0.697
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr4_-_54232007
|
0.696
|
NM_152540
|
SCFD2
|
sec1 family domain containing 2
|
|
chr20_+_33292117
|
0.695
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr5_+_50679239
|
0.693
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr6_+_41888993
|
0.692
|
|
BYSL
|
bystin-like
|
|
chr3_+_93698959
|
0.690
|
NM_001174150
NM_001174151
NM_144996
NM_182896
|
ARL13B
|
ADP-ribosylation factor-like 13B
|
|
chr10_+_22634373
|
0.680
|
NM_001253854
NM_001253855
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr20_+_18488167
|
0.679
|
NM_006363
NM_032985
NM_032986
NM_001172746
|
SEC23B
|
Sec23 homolog B (S. cerevisiae)
|
|
chr6_+_41888964
|
0.677
|
NM_004053
|
BYSL
|
bystin-like
|
|
chr7_+_6144589
|
0.675
|
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr3_-_195808958
|
0.675
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr1_+_87794150
|
0.673
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr11_-_3818706
|
0.672
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr3_-_154042204
|
0.668
|
NM_001114397
NM_020865
|
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36
|
|
chr3_-_185655776
|
0.666
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr19_+_3762671
|
0.664
|
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr3_+_186739643
|
0.662
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr6_+_64281910
|
0.659
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr3_-_186288331
|
0.658
|
NM_001134415
|
TBCCD1
|
TBCC domain containing 1
|
|
chr6_-_108395854
|
0.650
|
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr12_+_56660724
|
0.649
|
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr3_+_99536667
|
0.648
|
NM_032359
|
C3orf26
|
chromosome 3 open reading frame 26
|
|
chr7_-_8301681
|
0.647
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr12_+_56661026
|
0.647
|
NM_001099337
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr3_-_195808997
|
0.640
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr6_-_108395918
|
0.637
|
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr16_+_70148213
|
0.637
|
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr19_+_3762650
|
0.635
|
NM_172251
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr14_-_76448091
|
0.635
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr18_+_20513277
|
0.634
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr19_-_44100127
|
0.632
|
NM_001007561
|
IRGQ
|
immunity-related GTPase family, Q
|
|
chr16_+_475605
|
0.631
|
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chrX_+_44732402
|
0.630
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr7_+_22766765
|
0.628
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr17_-_47785527
|
0.626
|
|
SLC35B1
|
solute carrier family 35, member B1
|
|
chr17_-_47785132
|
0.625
|
|
SLC35B1
|
solute carrier family 35, member B1
|
|
chr8_+_64081187
|
0.623
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr9_-_70490123
|
0.622
|
|
CBWD3
|
COBW domain containing 3
|
|
chr8_-_74791078
|
0.620
|
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr1_-_31769599
|
0.619
|
NM_004814
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5)
|
|
chrX_-_13956644
|
0.619
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr17_-_8151353
|
0.618
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr6_-_31846751
|
0.618
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr12_+_59989847
|
0.617
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr20_+_42086468
|
0.615
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr1_+_231114839
|
0.613
|
|
|
|
|
chr17_+_2497233
|
0.613
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr8_-_29207795
|
0.611
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr12_+_56660548
|
0.610
|
NM_144576
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr19_-_14316980
|
0.608
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr6_+_43211221
|
0.606
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr16_+_475629
|
0.604
|
NM_014700
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr6_+_39760148
|
0.604
|
NM_001201427
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chrX_-_7895424
|
0.603
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr17_-_47785270
|
0.602
|
NM_005827
|
SLC35B1
|
solute carrier family 35, member B1
|
|
chr22_+_21996591
|
0.598
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr9_-_127703294
|
0.597
|
NM_002077
|
GOLGA1
|
golgin A1
|
|
chr9_+_91933384
|
0.596
|
NM_024077
|
SECISBP2
|
SECIS binding protein 2
|
|
chr1_+_218518675
|
0.595
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr3_-_185655648
|
0.595
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|
|
chr1_-_38273823
|
0.595
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr12_+_7282963
|
0.595
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr19_+_49375677
|
0.587
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr5_+_148206155
|
0.586
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr11_-_126138824
|
0.586
|
NM_001177842
NM_003139
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr8_+_17104384
|
0.586
|
NM_001145152
NM_152415
|
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae)
|
|
chr22_+_21996572
|
0.585
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr2_-_9563584
|
0.585
|
|
ITGB1BP1
|
integrin beta 1 binding protein 1
|
|
chr17_-_72968790
|
0.585
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr3_+_186288445
|
0.583
|
NM_016306
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11
|
|
chr11_-_126138736
|
0.582
|
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr7_+_22767101
|
0.580
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr19_+_49375679
|
0.578
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr5_+_50678957
|
0.578
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr8_-_71581354
|
0.576
|
|
LACTB2
|
lactamase, beta 2
|
|
chr8_-_74791072
|
0.575
|
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr8_+_123793894
|
0.574
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr19_-_18314731
|
0.573
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr1_-_11120001
|
0.571
|
|
SRM
|
spermidine synthase
|
|
chr11_+_65337923
|
0.569
|
NM_006396
|
SSSCA1
|
Sjogren syndrome/scleroderma autoantigen 1
|
|
chr3_-_185655806
|
0.567
|
|
TRA2B
|
transformer 2 beta homolog (Drosophila)
|