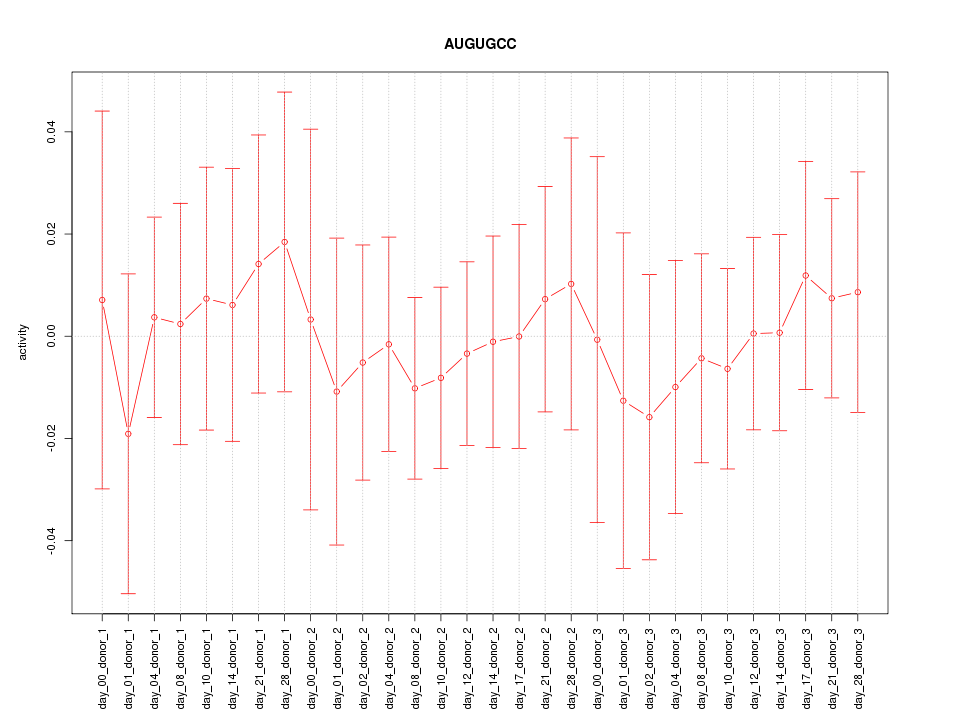
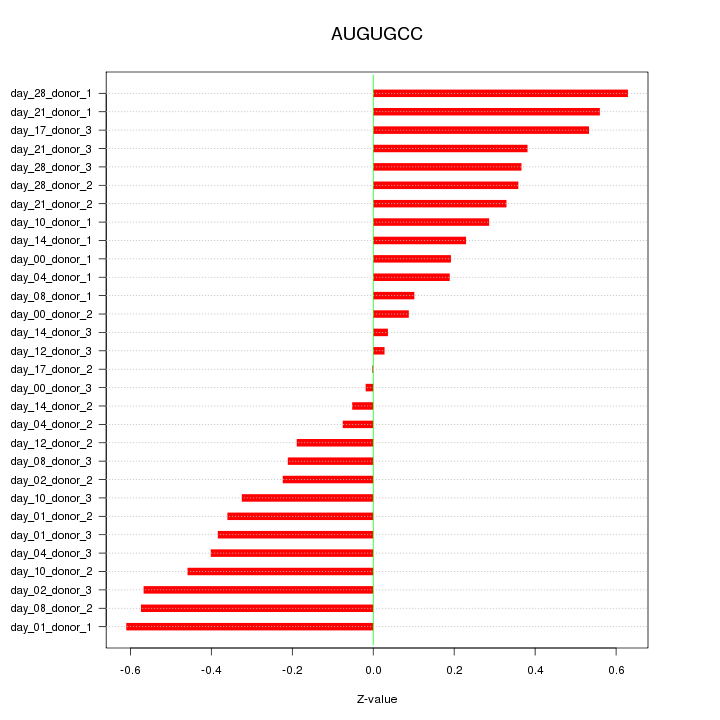
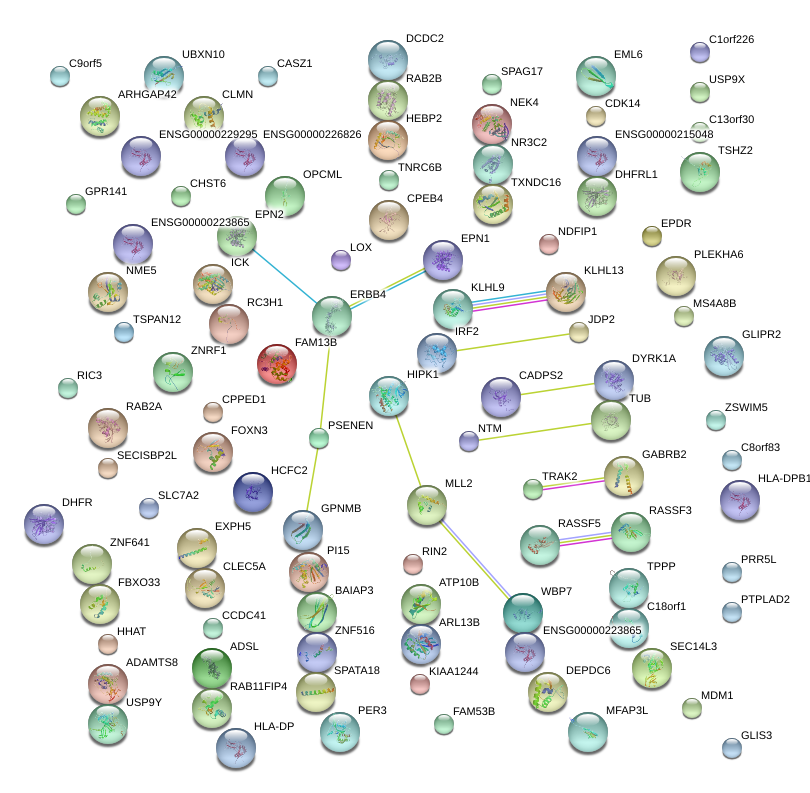
|
chr9_-_4152182
|
0.443
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr9_-_4300028
|
0.441
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr7_+_37960162
|
0.425
|
NM_001242946
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr13_+_43355685
|
0.395
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr4_-_149363498
|
0.380
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr11_+_60467046
|
0.356
|
NM_031457
|
MS4A8B
|
membrane-spanning 4-domains, subfamily A, member 8B
|
|
chr18_+_13218777
|
0.317
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr12_+_104458235
|
0.311
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr14_-_90085325
|
0.297
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr18_+_13611664
|
0.288
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr18_+_13620866
|
0.285
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr5_-_137475076
|
0.272
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr1_+_20512577
|
0.262
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr2_-_213403280
|
0.260
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr18_-_74207145
|
0.260
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr1_-_173962050
|
0.260
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr14_-_89883306
|
0.248
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr1_-_204329043
|
0.246
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr5_-_693413
|
0.220
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr4_+_52917440
|
0.216
|
NM_145263
|
SPATA18
|
spermatogenesis associated 18 homolog (rat)
|
|
chr8_+_17354562
|
0.215
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr14_-_95786095
|
0.214
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr11_-_8190535
|
0.198
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chr22_-_30867944
|
0.190
|
NM_174975
|
SEC14L3
|
SEC14-like 3 (S. cerevisiae)
|
|
chr12_-_68726036
|
0.187
|
NM_001205028
NM_001205029
NM_017440
NM_020128
|
MDM1
|
Mdm1 nuclear protein homolog (mouse)
|
|
chr8_+_17396285
|
0.176
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr14_-_39901619
|
0.176
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr16_-_75528925
|
0.175
|
NM_021615
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6
|
|
chr9_+_36136710
|
0.172
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr12_-_48744553
|
0.167
|
NM_001172681
NM_152320
|
ZNF641
|
zinc finger protein 641
|
|
chr6_+_33043702
|
0.167
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr5_-_121414011
|
0.166
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr20_-_18447622
|
0.165
|
NM_001099407
|
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1
|
|
chr5_-_121412805
|
0.160
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr12_-_48745020
|
0.158
|
NM_001172682
|
ZNF641
|
zinc finger protein 641
|
|
chr2_-_202316227
|
0.156
|
NM_015049
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr22_+_40440664
|
0.152
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr14_+_75894919
|
0.151
|
NM_001135048
NM_130469
|
JDP2
|
Jun dimerization protein 2
|
|
chr15_-_77712436
|
0.148
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr17_+_19140674
|
0.147
|
NM_001102664
NM_014964
NM_148921
|
EPN2
|
epsin 2
|
|
chr1_+_210502639
|
0.142
|
NM_001170580
|
HHAT
|
hedgehog acyltransferase
|
|
chr7_+_90338711
|
0.142
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr7_-_122526781
|
0.138
|
NM_001009571
NM_001167940
NM_017954
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr8_+_75736771
|
0.136
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr7_+_23286277
|
0.134
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr5_+_141488314
|
0.133
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr12_-_94853696
|
0.132
|
NM_001042399
NM_016122
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr9_-_21031607
|
0.130
|
NM_001010915
|
PTPLAD2
|
protein tyrosine phosphatase-like A domain containing 2
|
|
chr1_+_210501595
|
0.129
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr1_-_10856704
|
0.128
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr1_+_210502188
|
0.125
|
NM_001170587
NM_001170588
NM_018194
|
HHAT
|
hedgehog acyltransferase
|
|
chr21_+_38739858
|
0.124
|
NM_101395
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr8_-_93978264
|
0.123
|
NM_001191035
NM_001171795
NM_001171796
NM_001171797
NM_001171798
NM_001171799
|
C8orf83
|
chromosome 8 open reading frame 83
|
|
chr1_-_118727780
|
0.119
|
NM_206996
|
SPAG17
|
sperm associated antigen 17
|
|
chr16_-_12897693
|
0.119
|
NM_001099455
NM_018340
|
CPPED1
|
calcineurin-like phosphoesterase domain containing 1
|
|
chr14_+_75898836
|
0.119
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr20_+_51588945
|
0.118
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr4_-_170924911
|
0.118
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr16_+_1383605
|
0.115
|
NM_001199097
NM_001199098
NM_001199099
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr4_-_185395616
|
0.114
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr12_-_49449106
|
0.114
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr5_-_137368778
|
0.112
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr11_-_132812870
|
0.112
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chrX_-_117251302
|
0.112
|
NM_001168301
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr5_+_173315313
|
0.111
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr20_+_51801821
|
0.111
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chrX_-_117107700
|
0.107
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr16_+_1384504
|
0.107
|
NM_003933
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr20_+_19867075
|
0.107
|
NM_001242581
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_+_138483024
|
0.106
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr15_-_49338496
|
0.104
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chrX_-_117250624
|
0.103
|
NM_001168302
NM_001168303
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chrX_-_117119282
|
0.102
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr3_+_93698959
|
0.102
|
NM_001174150
NM_001174151
NM_144996
NM_182896
|
ARL13B
|
ADP-ribosylation factor-like 13B
|
|
chr11_+_36422546
|
0.101
|
NM_001160169
|
PRR5L
|
proline rich 5 like
|
|
chr1_+_162351519
|
0.098
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr12_+_65004191
|
0.097
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr20_+_19870209
|
0.097
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr5_-_160975124
|
0.094
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr1_+_114496498
|
0.094
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr4_-_170947428
|
0.094
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr8_+_120885899
|
0.093
|
NM_022783
|
DEPTOR
|
DEP domain containing MTOR-interacting protein
|
|
chr11_+_36397534
|
0.093
|
NM_001160168
NM_024841
|
PRR5L
|
proline rich 5 like
|
|
chr1_+_114493766
|
0.092
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr14_+_75894508
|
0.092
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr3_-_52804871
|
0.090
|
NM_001193533
NM_003157
|
NEK4
|
NIMA (never in mitosis gene a)-related kinase 4
|
|
chr17_+_29718641
|
0.088
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr1_+_7844713
|
0.087
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr16_+_1386197
|
0.084
|
NM_001199096
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr10_-_126432893
|
0.083
|
NM_014661
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr21_+_38792600
|
0.083
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr14_-_53019036
|
0.081
|
NM_001160047
NM_020784
|
TXNDC16
|
thioredoxin domain containing 16
|
|
chr11_+_36317724
|
0.081
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr6_-_52926588
|
0.080
|
NM_014920
NM_016513
|
ICK
|
intestinal cell (MAK-like) kinase
|
|
chr16_+_75032874
|
0.079
|
NM_032268
|
ZNRF1
|
zinc and ring finger 1
|
|
chr1_-_45672225
|
0.079
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr3_-_93782066
|
0.078
|
NM_001195643
|
DHFRL1
|
dihydrofolate reductase-like 1
|
|
chr7_-_120498128
|
0.078
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr9_-_111882209
|
0.078
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr11_+_100557851
|
0.077
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr19_+_36236460
|
0.077
|
NM_172341
|
PSENEN
|
presenilin enhancer 2 homolog (C. elegans)
|
|
chr5_-_160279045
|
0.076
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr6_-_24358279
|
0.076
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr7_-_141646716
|
0.075
|
NM_013252
|
CLEC5A
|
C-type lectin domain family 5, member A
|
|
chr3_-_93781659
|
0.074
|
NM_176815
|
DHFRL1
|
dihydrofolate reductase-like 1
|
|
chr2_+_54952148
|
0.074
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr8_+_61429390
|
0.073
|
NM_001242644
NM_002865
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr11_-_108464344
|
0.072
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr11_+_8102691
|
0.071
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chrX_+_40944858
|
0.070
|
NM_001039590
NM_001039591
|
USP9X
|
ubiquitin specific peptidase 9, X-linked
|
|
chr20_+_62184372
|
0.070
|
NM_024059
|
C20orf195
|
chromosome 20 open reading frame 195
|
|
chr1_-_207095198
|
0.068
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr9_-_126030854
|
0.067
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr6_-_24383519
|
0.067
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr17_-_80606095
|
0.066
|
NM_019613
|
WDR45L
|
WDR45-like
|
|
chr19_+_57702833
|
0.066
|
NM_003417
|
ZNF264
|
zinc finger protein 264
|
|
chr5_+_113698015
|
0.064
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr8_-_23540401
|
0.064
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr4_-_66535652
|
0.064
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr3_-_141944441
|
0.063
|
NM_001039547
|
GK5
|
glycerol kinase 5 (putative)
|
|
chr5_+_130506635
|
0.063
|
NM_181705
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr11_+_11862955
|
0.062
|
NM_017944
|
USP47
|
ubiquitin specific peptidase 47
|
|
chr14_-_67982020
|
0.062
|
NM_182526
|
TMEM229B
|
transmembrane protein 229B
|
|
chr2_-_240322620
|
0.062
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr9_-_126030792
|
0.062
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr5_+_36608430
|
0.062
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr11_+_43380403
|
0.061
|
NM_018259
|
TTC17
|
tetratricopeptide repeat domain 17
|
|
chr22_+_40573879
|
0.060
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr17_+_30264039
|
0.060
|
NM_015355
|
SUZ12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chr5_-_132073143
|
0.060
|
NM_007054
|
KIF3A
|
kinesin family member 3A
|
|
chr16_+_15489586
|
0.060
|
NM_001128423
NM_173803
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr5_+_113769226
|
0.060
|
NM_170775
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr10_+_45869611
|
0.059
|
NM_000698
|
ALOX5
|
arachidonate 5-lipoxygenase
|
|
chr16_-_27899477
|
0.059
|
NM_144675
|
GSG1L
|
GSG1-like
|
|
chr12_-_40499604
|
0.057
|
NM_052885
|
SLC2A13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr4_-_186697065
|
0.057
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr15_+_101459459
|
0.056
|
NM_024652
|
LRRK1
|
leucine-rich repeat kinase 1
|
|
chr5_+_38258510
|
0.056
|
NM_001205301
NM_152403
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr2_+_63277191
|
0.056
|
NM_001199770
|
OTX1
|
orthodenticle homeobox 1
|
|
chr4_-_186877869
|
0.056
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_+_2162388
|
0.056
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr2_-_38303307
|
0.055
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr20_-_44880324
|
0.055
|
NM_021248
|
CDH22
|
cadherin 22, type 2
|
|
chr12_+_65218351
|
0.055
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr17_+_57642885
|
0.054
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chrX_+_67718623
|
0.054
|
NM_001195214
NM_173834
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr10_+_24497719
|
0.053
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr10_-_93392857
|
0.053
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr20_+_13976014
|
0.053
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr14_+_77648082
|
0.052
|
NM_020431
|
TMEM63C
|
transmembrane protein 63C
|
|
chr8_+_42396716
|
0.052
|
NM_138436
NM_001135674
NM_001135675
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr1_+_36772696
|
0.052
|
NM_024676
|
SH3D21
|
SH3 domain containing 21
|
|
chr17_-_40264687
|
0.052
|
NM_024119
|
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58
|
|
chr8_+_8559665
|
0.052
|
NM_194284
|
CLDN23
|
claudin 23
|
|
chrX_+_144908927
|
0.052
|
NM_004709
|
CXorf1
|
chromosome X open reading frame 1
|
|
chr20_+_15177503
|
0.052
|
NM_001033087
|
MACROD2
|
MACRO domain containing 2
|
|
chr19_-_33166057
|
0.051
|
NM_032139
|
ANKRD27
|
ankyrin repeat domain 27 (VPS9 domain)
|
|
chr8_+_42396297
|
0.050
|
NM_001135676
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr2_+_63277936
|
0.050
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr8_-_61193882
|
0.050
|
NM_004056
|
CA8
|
carbonic anhydrase VIII
|
|
chr4_-_85887543
|
0.050
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chrX_+_77166178
|
0.050
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr5_+_102465256
|
0.049
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr11_-_74660165
|
0.049
|
NM_182969
|
XRRA1
|
X-ray radiation resistance associated 1
|
|
chr7_-_82792127
|
0.049
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr16_-_28074776
|
0.049
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr5_+_38445577
|
0.049
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr1_+_32716839
|
0.049
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr2_-_152955502
|
0.049
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chrX_+_37865834
|
0.048
|
NM_001163335
NM_138780
|
SYTL5
|
synaptotagmin-like 5
|
|
chr14_-_69619870
|
0.048
|
NM_003861
|
DCAF5
|
DDB1 and CUL4 associated factor 5
|
|
chr16_+_14013989
|
0.048
|
NM_005236
|
ERCC4
|
excision repair cross-complementing rodent repair deficiency, complementation group 4
|
|
chr21_+_38791206
|
0.048
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr7_-_64451413
|
0.048
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chr3_+_183353355
|
0.048
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr12_+_12869818
|
0.047
|
NM_004064
|
CDKN1B
|
cyclin-dependent kinase inhibitor 1B (p27, Kip1)
|
|
chr3_-_69101236
|
0.047
|
NM_007114
|
TMF1
|
TATA element modulatory factor 1
|
|
chr7_+_70597788
|
0.047
|
NM_022479
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17
|
|
chr9_-_5833072
|
0.047
|
NM_024896
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1
|
|
chr2_+_85198176
|
0.047
|
NM_020122
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chr2_-_152955208
|
0.047
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr16_-_31021716
|
0.046
|
NM_052874
|
STX1B
|
syntaxin 1B
|
|
chr14_+_96671015
|
0.046
|
NM_000623
|
BDKRB2
|
bradykinin receptor B2
|
|
chr1_+_212475147
|
0.046
|
NM_001199756
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr4_-_186732047
|
0.046
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr18_+_51795848
|
0.045
|
NM_007195
|
POLI
|
polymerase (DNA directed) iota
|
|
chr10_+_23983674
|
0.045
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr3_+_108541544
|
0.044
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr4_-_186578122
|
0.044
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_48136245
|
0.044
|
NM_003328
|
TXK
|
TXK tyrosine kinase
|
|
chr5_+_110559647
|
0.043
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr2_-_175351755
|
0.043
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr17_+_26698986
|
0.043
|
NM_015077
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chrX_+_152801579
|
0.043
|
NM_001001344
NM_021949
|
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3
|
|
chrX_-_67653169
|
0.043
|
NM_002547
|
OPHN1
|
oligophrenin 1
|
|
chrX_+_37892786
|
0.042
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr9_+_120466453
|
0.042
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chrX_-_20135015
|
0.042
|
NM_001168465
NM_001168466
NM_152780
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr12_+_52345437
|
0.042
|
NM_004302
NM_020328
|
ACVR1B
|
activin A receptor, type IB
|
|
chr8_-_101322245
|
0.042
|
NM_183419
|
RNF19A
|
ring finger protein 19A
|