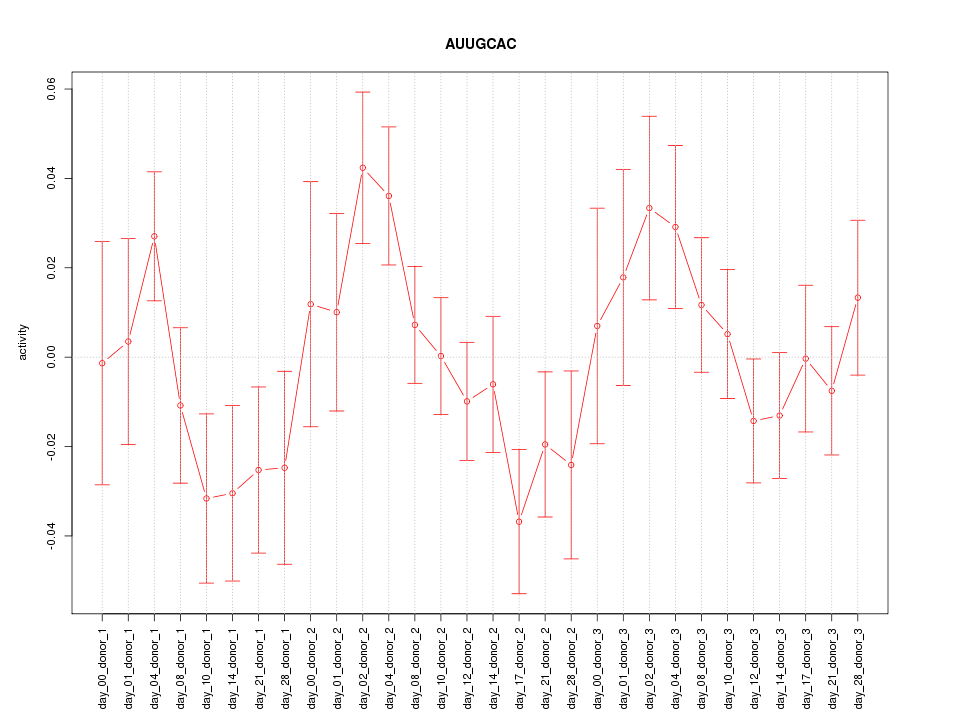
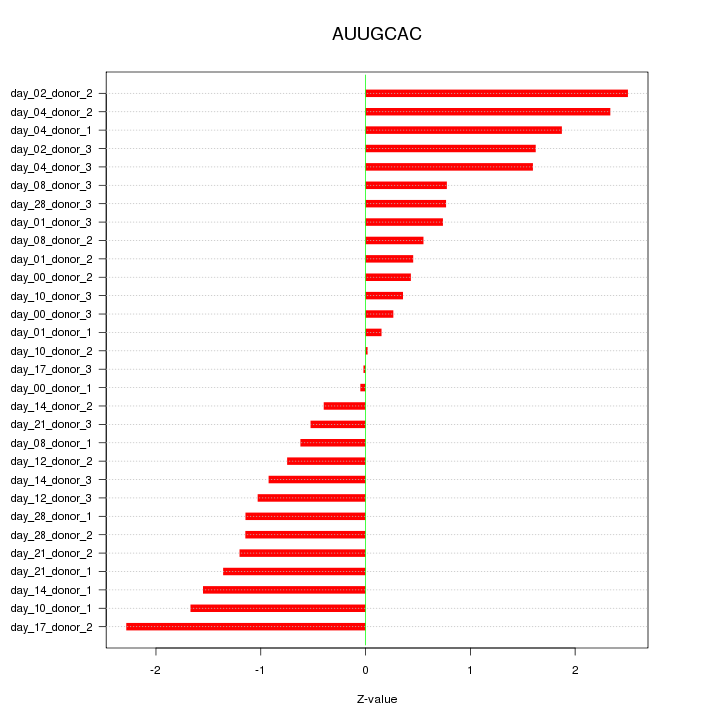
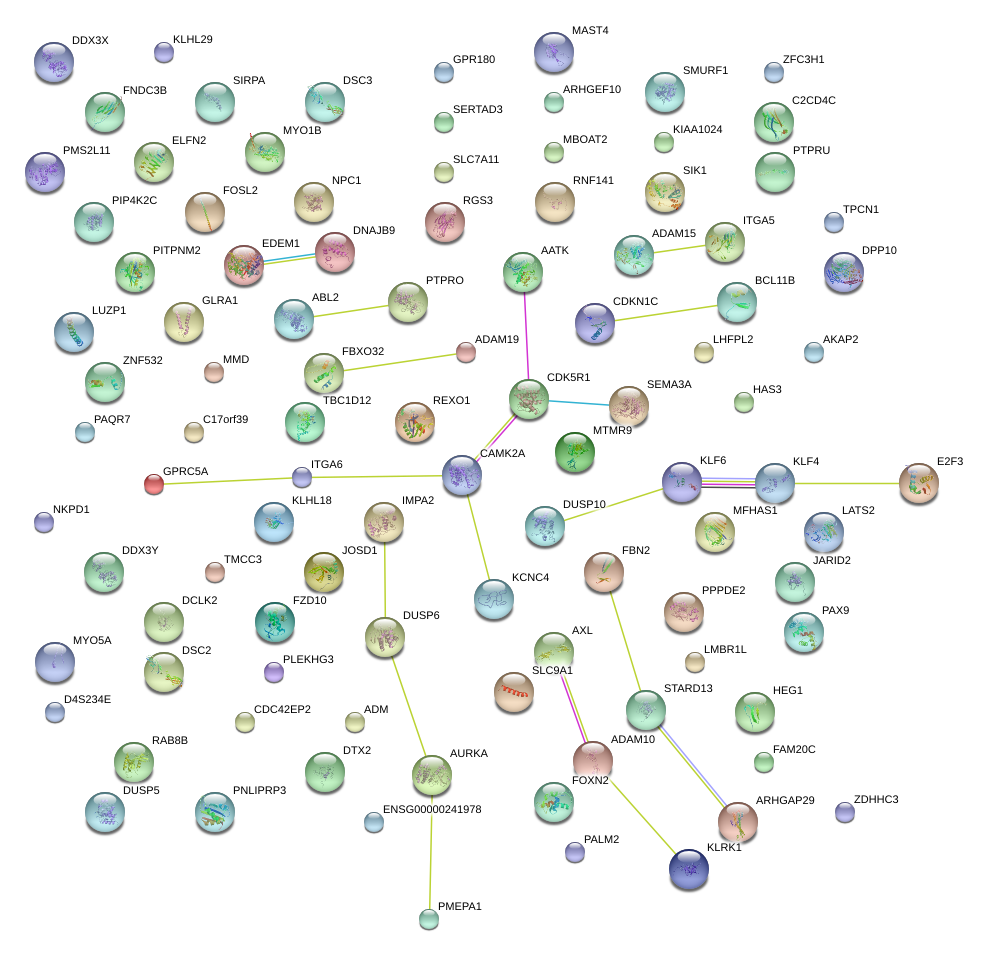
|
chr20_-_56286540
|
3.001
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr21_-_44846927
|
2.803
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr20_-_56284945
|
2.774
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr2_+_23608297
|
2.668
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr16_+_69139466
|
2.667
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr5_-_149669400
|
2.651
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr16_+_69141442
|
2.544
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr20_-_56285576
|
2.497
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr4_-_139163222
|
2.413
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr18_-_28681885
|
2.316
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr5_-_157002739
|
2.276
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr12_-_95044205
|
1.724
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr18_-_21166440
|
1.616
|
NM_000271
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr19_+_41725098
|
1.616
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr18_+_56530060
|
1.588
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr12_-_54813010
|
1.563
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr10_+_118187381
|
1.430
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr5_-_77944341
|
1.421
|
NM_005779
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2
|
|
chr12_+_15475190
|
1.413
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr22_-_37823503
|
1.397
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr9_+_116356199
|
1.382
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_116207010
|
1.375
|
NM_144488
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_+_48541767
|
1.371
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr6_+_20402040
|
1.337
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr11_-_2906960
|
1.332
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr14_+_65171149
|
1.320
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr10_+_112257624
|
1.315
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr12_+_15699277
|
1.280
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr1_-_26197743
|
1.270
|
NM_178422
|
PAQR7
|
progestin and adipoQ receptor family member VII
|
|
chr20_-_54967221
|
1.245
|
NM_003600
NM_198433
NM_198434
NM_198435
NM_198436
NM_198437
|
AURKA
|
aurora kinase A
|
|
chr3_-_124774735
|
1.243
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr12_-_89746244
|
1.228
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr7_-_98741681
|
1.223
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr10_-_3827418
|
1.218
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr8_-_8750739
|
1.218
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr11_+_65082288
|
1.211
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr9_+_116342939
|
1.201
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_221915458
|
1.179
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_+_110753313
|
1.176
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr9_+_116263706
|
1.155
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_116327301
|
1.145
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr15_+_63481713
|
1.130
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr1_-_221910801
|
1.130
|
NM_144728
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr3_+_47324329
|
1.112
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr2_+_173292306
|
1.109
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr20_+_1875382
|
1.097
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr20_+_1875825
|
1.090
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr8_-_124544376
|
1.086
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr7_-_83824216
|
1.081
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr6_+_15246299
|
1.053
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr17_-_53499055
|
1.047
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr20_+_1874779
|
1.024
|
NM_001040022
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr2_+_192109879
|
0.994
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr12_+_13043955
|
0.986
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr7_+_76090971
|
0.984
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr15_+_79724857
|
0.983
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr12_-_123594974
|
0.976
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr5_-_151304322
|
0.969
|
NM_000171
NM_001146040
|
GLRA1
|
glycine receptor, alpha 1
|
|
chr4_+_4387982
|
0.956
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr7_+_108210188
|
0.943
|
NM_012328
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr2_-_9143742
|
0.936
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr2_+_28615771
|
0.931
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_115822246
|
0.930
|
NM_001178037
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr11_-_10562689
|
0.930
|
NM_016422
|
RNF141
|
ring finger protein 141
|
|
chr13_-_33859835
|
0.930
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_+_130647002
|
0.923
|
NM_007197
|
FZD10
|
frizzled family receptor 10
|
|
chr8_-_124553448
|
0.908
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr15_-_52821004
|
0.891
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr1_-_94703252
|
0.891
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_-_45017600
|
0.866
|
NM_001135179
NM_016598
|
ZDHHC3
|
zinc finger, DHHC-type containing 3
|
|
chr15_-_59041933
|
0.847
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr12_+_113659216
|
0.844
|
NM_001143819
NM_017901
|
TPCN1
|
two pore segment channel 1
|
|
chr14_+_37126772
|
0.833
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr1_-_27481400
|
0.827
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr17_+_30813928
|
0.821
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr12_-_72057676
|
0.821
|
NM_144982
|
ZFC3H1
|
zinc finger, C3H1-type containing
|
|
chr2_+_115199898
|
0.821
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr19_-_40950238
|
0.815
|
NM_203344
|
SERTAD3
|
SERTA domain containing 3
|
|
chr1_-_23495347
|
0.813
|
NM_001142546
NM_033631
|
LUZP1
|
leucine zipper protein 1
|
|
chr22_-_42017032
|
0.809
|
NM_015704
|
PPPDE2
|
PPPDE peptidase domain containing 2
|
|
chr10_+_96162185
|
0.804
|
NM_015188
|
TBC1D12
|
TBC1 domain family, member 12
|
|
chr13_-_21635703
|
0.801
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr14_-_99737821
|
0.797
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr1_+_155023741
|
0.796
|
NM_003815
NM_207191
NM_207194
NM_207195
NM_207196
NM_207197
|
ADAM15
|
ADAM metallopeptidase domain 15
|
|
chr5_-_127873734
|
0.794
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr17_+_17942584
|
0.789
|
NM_024052
|
C17orf39
|
chromosome 17 open reading frame 39
|
|
chr3_+_171757413
|
0.786
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr5_+_66124603
|
0.786
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr19_-_409169
|
0.782
|
NM_001136263
|
C2CD4C
|
C2 calcium-dependent domain containing 4C
|
|
chr13_-_33760215
|
0.781
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_+_76109826
|
0.773
|
NM_001102596
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr7_+_192968
|
0.770
|
NM_020223
|
FAM20C
|
family with sequence similarity 20, member C
|
|
chr9_-_110252045
|
0.770
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr3_+_5229357
|
0.764
|
NM_014674
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr3_+_171758289
|
0.750
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr13_-_33780142
|
0.749
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chrX_+_41192650
|
0.733
|
NM_001193416
NM_001193417
NM_001356
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked
|
|
chr17_-_79105747
|
0.729
|
NM_004920
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr19_-_1848447
|
0.725
|
NM_020695
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)
|
|
chr22_-_39095991
|
0.721
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr12_+_57984914
|
0.713
|
NM_001146258
NM_001146259
NM_001146260
NM_024779
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr2_+_115219153
|
0.698
|
NM_001178036
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr1_-_179198814
|
0.697
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr18_+_11981421
|
0.695
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr17_-_79139871
|
0.691
|
NM_001080395
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr4_+_150999412
|
0.689
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr9_+_112403067
|
0.688
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr11_+_10326618
|
0.686
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr13_+_95254021
|
0.684
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr8_+_1772095
|
0.683
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr8_+_11141983
|
0.683
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr19_-_45663407
|
0.682
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr12_-_49504646
|
0.673
|
NM_018113
|
LMBR1L
|
limb region 1 homolog (mouse)-like
|
|
chr3_-_48594209
|
0.671
|
NM_004567
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr6_+_70576447
|
0.670
|
NM_001858
|
COL19A1
|
collagen, type XIX, alpha 1
|
|
chr16_-_89007605
|
0.670
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chrX_+_47077426
|
0.668
|
NM_033018
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr19_+_47104490
|
0.665
|
NM_005184
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr22_-_29075708
|
0.663
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr16_+_11762235
|
0.659
|
NM_003498
|
SNN
|
stannin
|
|
chr3_+_38206968
|
0.657
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr2_-_25141855
|
0.651
|
NM_004036
|
ADCY3
|
adenylate cyclase 3
|
|
chr10_+_120967094
|
0.647
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr6_-_10415438
|
0.644
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr18_+_19749403
|
0.641
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr5_-_132299263
|
0.636
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr1_+_218518675
|
0.631
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr4_-_153303663
|
0.631
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr8_+_23101149
|
0.625
|
NM_152272
|
CHMP7
|
charged multivesicular body protein 7
|
|
chr5_-_179780314
|
0.623
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr12_-_80307690
|
0.618
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr6_-_82957441
|
0.615
|
NM_015525
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase
|
|
chr2_-_107502609
|
0.614
|
NM_001142351
|
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2
|
|
chr10_+_74033676
|
0.609
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr11_-_12030128
|
0.608
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr1_+_101702304
|
0.606
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr9_+_5629110
|
0.598
|
NM_001135920
NM_001206557
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr14_-_35182944
|
0.590
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr2_-_46384
|
0.588
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr18_+_3451589
|
0.585
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_-_38471170
|
0.578
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr9_-_140353785
|
0.573
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr9_+_124329398
|
0.573
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr5_+_72251753
|
0.566
|
NM_001146032
NM_138782
|
FCHO2
|
FCH domain only 2
|
|
chr14_-_35183722
|
0.562
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr19_-_40948504
|
0.558
|
NM_013368
|
SERTAD3
|
SERTA domain containing 3
|
|
chr1_+_10270680
|
0.556
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr3_-_120169493
|
0.556
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr6_-_80656870
|
0.554
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr3_-_113465101
|
0.550
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr11_+_13690151
|
0.550
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr17_-_42580927
|
0.549
|
NM_001002909
|
GPATCH8
|
G patch domain containing 8
|
|
chr10_-_105614952
|
0.549
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr6_-_10419786
|
0.545
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr8_+_58906968
|
0.544
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr11_-_12030622
|
0.543
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr1_-_193155723
|
0.542
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr11_-_12030882
|
0.538
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chrX_-_41782230
|
0.537
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr22_-_36784055
|
0.535
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr15_-_49447765
|
0.535
|
NM_001143887
NM_004236
|
COPS2
|
COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis)
|
|
chr11_-_47788839
|
0.533
|
NM_015308
|
FNBP4
|
formin binding protein 4
|
|
chr1_-_179112178
|
0.527
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr5_+_109025135
|
0.525
|
NM_002372
|
MAN2A1
|
mannosidase, alpha, class 2A, member 1
|
|
chr7_-_106301329
|
0.524
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr2_+_187464931
|
0.522
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr11_+_122526339
|
0.521
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr12_-_80329234
|
0.517
|
NM_001143885
NM_001244990
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr8_+_24772454
|
0.515
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr6_+_163835668
|
0.512
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr11_-_89224413
|
0.506
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr2_+_115919512
|
0.505
|
NM_001178034
NM_001004360
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr2_+_5832778
|
0.504
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr12_-_80328926
|
0.503
|
NM_002480
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr6_+_21593963
|
0.498
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr17_+_48423382
|
0.495
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr2_+_203241044
|
0.494
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr2_+_10091791
|
0.494
|
NM_198182
|
GRHL1
|
grainyhead-like 1 (Drosophila)
|
|
chr12_+_56862300
|
0.493
|
NM_207344
|
SPRYD4
|
SPRY domain containing 4
|
|
chr6_-_153452383
|
0.489
|
NM_012419
|
RGS17
|
regulator of G-protein signaling 17
|
|
chr2_-_2334887
|
0.487
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr11_-_128457452
|
0.481
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_-_214724974
|
0.479
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chrX_-_37706816
|
0.479
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr4_-_176923572
|
0.476
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr14_+_92788924
|
0.475
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr3_-_127541975
|
0.473
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr10_-_115934363
|
0.473
|
NM_018017
|
C10orf118
|
chromosome 10 open reading frame 118
|
|
chr3_-_37217716
|
0.472
|
NM_001134369
NM_006309
NM_017724
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2
|
|
chr7_+_77469446
|
0.470
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr7_-_71877359
|
0.469
|
NM_031468
|
CALN1
|
calneuron 1
|
|
chr22_-_36424397
|
0.468
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chrX_+_47078053
|
0.468
|
NM_006201
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr10_+_99186017
|
0.467
|
NM_002629
|
PGAM1
|
phosphoglycerate mutase 1 (brain)
|
|
chr13_-_78493902
|
0.465
|
NM_001201397
|
EDNRB
|
endothelin receptor type B
|
|
chr7_-_92463211
|
0.463
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr20_-_25566137
|
0.462
|
NM_025176
|
NINL
|
ninein-like
|
|
chr11_-_119599253
|
0.460
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr19_-_3700466
|
0.454
|
NM_001195733
NM_012398
|
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma
|
|
chr4_+_699529
|
0.453
|
NM_006315
|
PCGF3
|
polycomb group ring finger 3
|