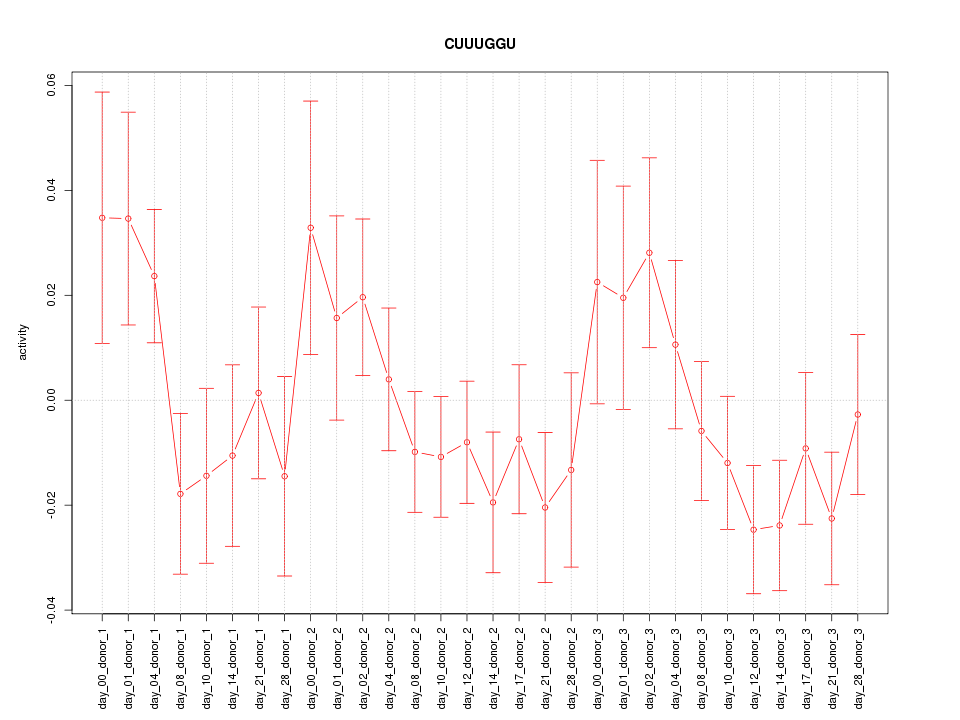
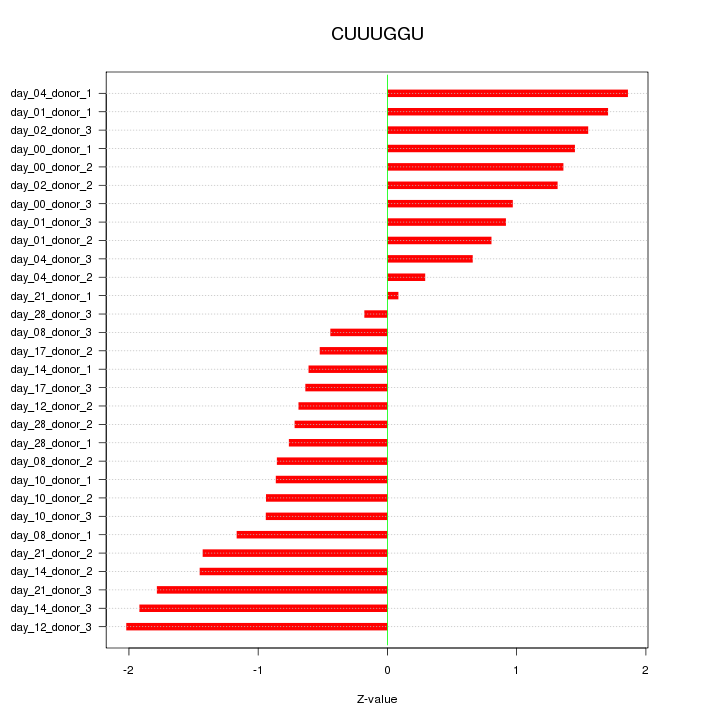
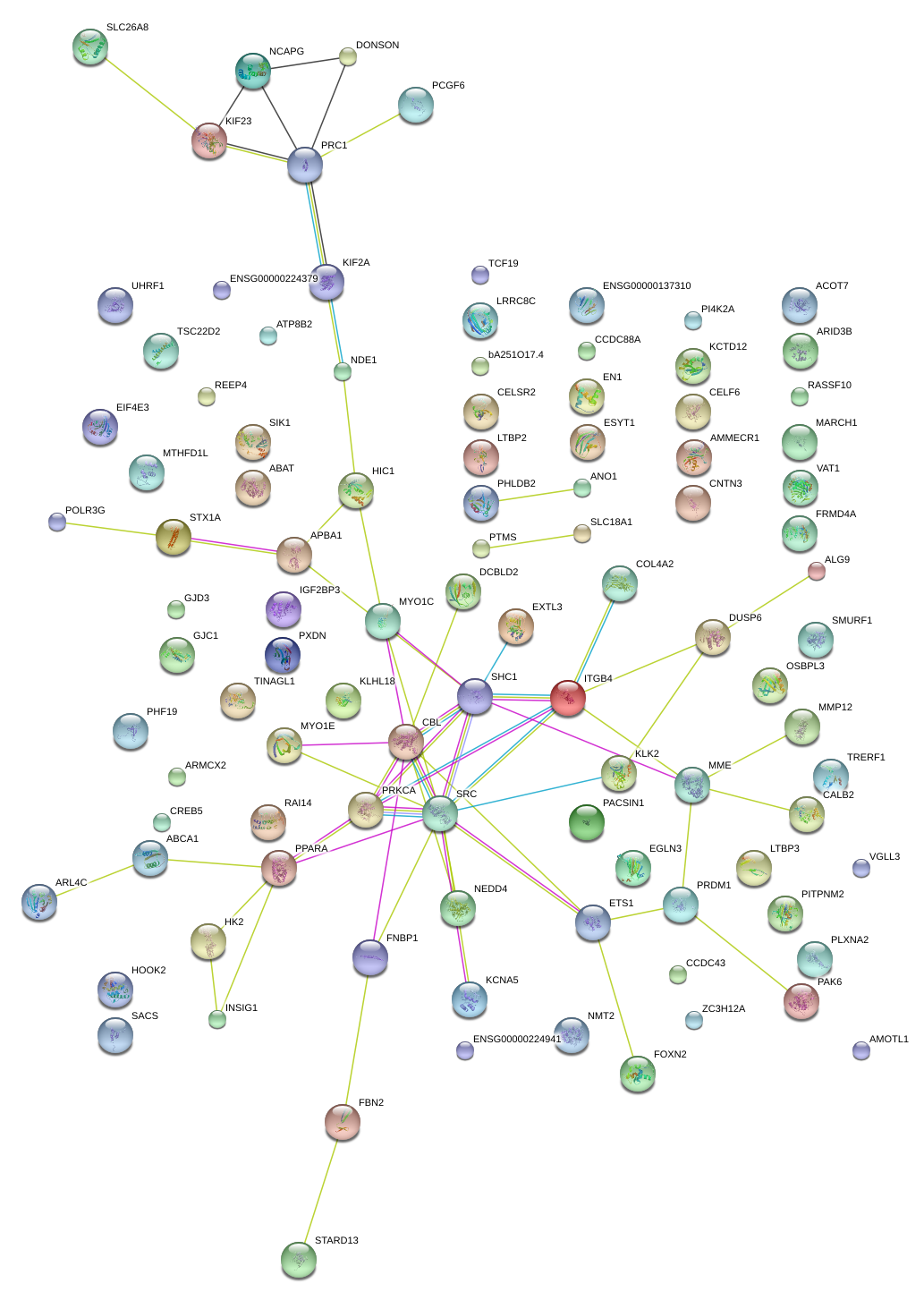
|
chr11_+_69924407
|
3.621
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr13_+_110959592
|
3.116
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr2_-_1748285
|
3.024
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr19_+_4910339
|
2.968
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr12_-_89746244
|
2.771
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr15_-_91537724
|
2.699
|
NM_003981
NM_199413
NM_199414
|
PRC1
|
protein regulator of cytokinesis 1
|
|
chr10_+_99400441
|
2.638
|
NM_018425
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr1_+_32042085
|
2.614
|
NM_001204414
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chrX_-_100914816
|
2.554
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr1_+_32043745
|
2.533
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr19_+_4909447
|
2.324
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr4_+_17812524
|
2.151
|
NM_022346
|
NCAPG
|
non-SMC condensin I complex, subunit G
|
|
chr11_+_94501503
|
2.133
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr1_-_6418966
|
2.021
|
NM_181866
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr9_-_72287097
|
1.964
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr6_+_106534188
|
1.955
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr15_+_69706619
|
1.935
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chr1_+_37940074
|
1.882
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr2_-_235405692
|
1.874
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr1_-_154946859
|
1.872
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr21_-_44846927
|
1.852
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr2_+_48541767
|
1.848
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr3_+_154797435
|
1.835
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr10_-_15210610
|
1.806
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr7_-_73133917
|
1.776
|
NM_001165903
NM_004603
|
STX1A
|
syntaxin 1A (brain)
|
|
chr7_-_98741681
|
1.756
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr1_-_6420719
|
1.754
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr11_-_128457452
|
1.743
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_-_6453773
|
1.714
|
NM_007274
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr1_+_90098643
|
1.705
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr17_-_41174431
|
1.703
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr5_-_127873734
|
1.694
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr6_+_31126114
|
1.671
|
NM_001077511
NM_007109
|
TCF19
|
transcription factor 19
|
|
chr1_-_6445737
|
1.607
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr1_-_154943001
|
1.603
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr11_-_128392061
|
1.599
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr16_+_15737123
|
1.566
|
NM_001143979
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr3_+_154798078
|
1.529
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_+_154797910
|
1.521
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr9_-_107690526
|
1.520
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr10_-_14372807
|
1.518
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr6_+_151187310
|
1.511
|
NM_001242768
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr2_+_75059781
|
1.495
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr7_-_23509972
|
1.488
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr21_-_34960937
|
1.484
|
NM_017613
|
DONSON
|
downstream neighbor of SON
|
|
chr6_-_35992269
|
1.480
|
NM_138718
NM_001193476
NM_052961
|
SLC26A8
|
solute carrier family 26, member 8
|
|
chr3_-_98620452
|
1.466
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr6_+_106546736
|
1.443
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr1_-_208417635
|
1.421
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr3_+_154797644
|
1.410
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr9_-_123639566
|
1.408
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr15_+_74833517
|
1.404
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr17_+_1958354
|
1.387
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr2_-_55646946
|
1.372
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr13_-_24007822
|
1.349
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr6_-_42419782
|
1.340
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr8_-_21998994
|
1.332
|
NM_025232
|
REEP4
|
receptor accessory protein 4
|
|
chrX_-_109683457
|
1.330
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr2_-_119605758
|
1.327
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr17_+_1959512
|
1.308
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr17_-_42907606
|
1.300
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr22_+_46546498
|
1.283
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr12_+_56521953
|
1.281
|
NM_001184796
NM_015292
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr8_+_28559143
|
1.281
|
NM_001440
|
EXTL3
|
exostoses (multiple)-like 3
|
|
chr13_-_33859835
|
1.268
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr16_+_71392615
|
1.258
|
NM_001740
NM_007088
|
CALB2
|
calbindin 2
|
|
chrX_-_109561314
|
1.238
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr3_+_47324329
|
1.231
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr15_-_56209305
|
1.230
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr16_+_15744082
|
1.216
|
NM_017668
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr11_+_119076974
|
1.205
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr3_-_71778268
|
1.187
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr14_-_75078669
|
1.180
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr2_+_64681270
|
1.172
|
NM_014181
|
LGALSL
|
lectin, galactoside-binding-like
|
|
chr17_-_1394849
|
1.167
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr16_+_8806825
|
1.141
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr17_-_42767016
|
1.129
|
NM_001099225
NM_144609
|
CCDC43
|
coiled-coil domain containing 43
|
|
chr7_+_28338939
|
1.118
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_+_111451326
|
1.113
|
NM_001134437
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr15_-_72598655
|
1.095
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr15_-_56285732
|
1.092
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr6_+_34482646
|
1.079
|
NM_001199583
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr3_+_111578026
|
1.077
|
NM_001134438
NM_145753
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr7_+_28725597
|
1.060
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr13_-_77460432
|
1.057
|
NM_138444
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr1_+_154300275
|
1.055
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr13_-_33760215
|
1.053
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr6_+_151186814
|
1.048
|
NM_001242767
NM_001242769
NM_015440
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr17_-_42908178
|
1.047
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr17_-_1389013
|
1.045
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr13_-_33780142
|
1.041
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr10_-_105110890
|
1.037
|
NM_001011663
NM_032154
|
PCGF6
|
polycomb group ring finger 6
|
|
chr12_-_123594974
|
1.036
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr16_+_8814567
|
1.034
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr11_-_111741980
|
1.025
|
NM_001077691
NM_001077692
|
ALG9
|
asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae)
|
|
chr5_+_34684611
|
1.014
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr17_+_64298844
|
0.978
|
NM_002737
|
PRKCA
|
protein kinase C, alpha
|
|
chr17_-_1395950
|
0.976
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr15_+_40509628
|
0.974
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr3_-_87040246
|
0.973
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr7_+_155089483
|
0.972
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr3_+_150126705
|
0.966
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr7_-_25019625
|
0.965
|
NM_015550
NM_145320
NM_145321
NM_145322
|
OSBPL3
|
oxysterol binding protein-like 3
|
|
chr14_-_34419957
|
0.962
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr17_+_73717515
|
0.960
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr5_+_89770680
|
0.951
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr20_+_35973083
|
0.936
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr9_-_132805456
|
0.935
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr1_+_109792640
|
0.934
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr11_+_13030879
|
0.924
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr4_-_165304406
|
0.922
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr3_-_74570262
|
0.922
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chrX_+_79675945
|
0.917
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr11_-_111742205
|
0.914
|
NM_001077690
NM_024740
|
ALG9
|
asparagine-linked glycosylation 9, alpha-1,2-mannosyltransferase homolog (S. cerevisiae)
|
|
chr1_-_155948317
|
0.911
|
NM_001162383
NM_001162384
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr8_-_28243941
|
0.908
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr2_+_191745546
|
0.905
|
NM_014905
|
GLS
|
glutaminase
|
|
chr5_+_135364583
|
0.901
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr17_-_49198084
|
0.901
|
NM_001130527
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr7_+_28475233
|
0.897
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_+_101419169
|
0.887
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr1_-_6321034
|
0.884
|
NM_207370
|
GPR153
|
G protein-coupled receptor 153
|
|
chr22_+_38302085
|
0.880
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr12_-_113909876
|
0.878
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr3_-_195808958
|
0.875
|
NM_003234
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr17_+_25621105
|
0.874
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr22_-_36424397
|
0.867
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr8_+_38089165
|
0.866
|
NM_001164232
|
DDHD2
|
DDHD domain containing 2
|
|
chr2_+_173292306
|
0.865
|
NM_000210
NM_001079818
|
ITGA6
|
integrin, alpha 6
|
|
chr7_+_28452143
|
0.859
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr13_+_98086315
|
0.855
|
NM_021033
|
RAP2A
|
RAP2A, member of RAS oncogene family
|
|
chr1_+_7831326
|
0.853
|
NM_004781
|
VAMP3
|
vesicle-associated membrane protein 3 (cellubrevin)
|
|
chr9_+_131102786
|
0.852
|
NM_005094
|
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr15_+_59063389
|
0.851
|
NM_001040450
NM_001040453
|
FAM63B
|
family with sequence similarity 63, member B
|
|
chr22_-_36784055
|
0.843
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr2_-_61245363
|
0.831
|
NM_144709
|
PUS10
|
pseudouridylate synthase 10
|
|
chr11_+_33563772
|
0.825
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr4_-_164534656
|
0.822
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr3_-_195808997
|
0.821
|
NM_001128148
|
TFRC
|
transferrin receptor (p90, CD71)
|
|
chr17_-_27044728
|
0.820
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr1_-_241803203
|
0.817
|
NM_014322
|
OPN3
|
opsin 3
|
|
chr6_-_80656870
|
0.814
|
NM_022726
|
ELOVL4
|
ELOVL fatty acid elongase 4
|
|
chr4_+_81187741
|
0.814
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr8_+_42752019
|
0.810
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr11_-_87908551
|
0.807
|
NM_022337
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr1_-_179198814
|
0.807
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr9_+_101705710
|
0.799
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr11_+_118477212
|
0.797
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr2_+_177053306
|
0.794
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr1_+_107682628
|
0.792
|
NM_001113226
NM_001113228
NM_014917
|
NTNG1
|
netrin G1
|
|
chr1_+_10270680
|
0.786
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr19_+_41222915
|
0.783
|
NM_025194
|
ITPKC
|
inositol-trisphosphate 3-kinase C
|
|
chr15_+_81293286
|
0.779
|
NM_022566
|
MESDC1
|
mesoderm development candidate 1
|
|
chr15_+_91411884
|
0.777
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr21_-_15755439
|
0.775
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr5_+_34687663
|
0.775
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr18_+_52495618
|
0.773
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr1_-_26233367
|
0.772
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr5_+_34656595
|
0.769
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr6_+_34433849
|
0.768
|
NM_020804
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr2_+_228336877
|
0.767
|
NM_001135187
NM_001135188
NM_001135189
NM_004504
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr2_-_69870976
|
0.757
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr3_-_123135188
|
0.754
|
NM_001199642
|
ADCY5
|
adenylate cyclase 5
|
|
chr7_+_100136784
|
0.751
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr20_+_35974458
|
0.751
|
NM_198291
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr3_+_38206968
|
0.745
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chrX_-_15873094
|
0.745
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr5_+_34656367
|
0.744
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr2_-_197791339
|
0.737
|
NM_024989
|
PGAP1
|
post-GPI attachment to proteins 1
|
|
chr3_+_101498285
|
0.734
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr1_+_155108276
|
0.733
|
NM_001122837
NM_001122839
NM_018845
|
SLC50A1
|
solute carrier family 50 (sugar transporter), member 1
|
|
chr12_-_109915127
|
0.726
|
NM_031954
|
KCTD10
|
potassium channel tetramerisation domain containing 10
|
|
chr8_+_22224997
|
0.723
|
NM_001135153
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14
|
|
chr3_-_185542807
|
0.722
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr13_+_46039024
|
0.720
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr1_-_155947898
|
0.717
|
NM_004723
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr6_-_138893666
|
0.717
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chrX_+_79591002
|
0.716
|
NM_001170574
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chrX_+_9754465
|
0.711
|
NM_001649
|
SHROOM2
|
shroom family member 2
|
|
chr14_+_53019865
|
0.711
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr1_-_27481400
|
0.710
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr18_+_29027731
|
0.709
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr5_-_64064483
|
0.708
|
NM_173829
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chr19_-_14224979
|
0.708
|
NM_207518
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr9_+_116638544
|
0.705
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr20_-_6104056
|
0.704
|
NM_017671
|
FERMT1
|
fermitin family member 1
|
|
chr2_-_110371669
|
0.701
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr6_+_116692098
|
0.701
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr6_-_138820578
|
0.701
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr1_-_26232643
|
0.697
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr22_-_36236421
|
0.694
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr3_+_101498028
|
0.691
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr8_+_1449531
|
0.690
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr11_+_118478305
|
0.686
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr10_+_102756829
|
0.684
|
NM_032429
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr6_+_35310317
|
0.681
|
NM_001171818
NM_001171819
NM_001171820
NM_006238
NM_177435
|
PPARD
|
peroxisome proliferator-activated receptor delta
|
|
chr7_+_77469446
|
0.681
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr11_+_122526339
|
0.679
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr1_+_154377668
|
0.676
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr14_-_48144156
|
0.675
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|