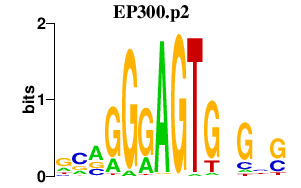
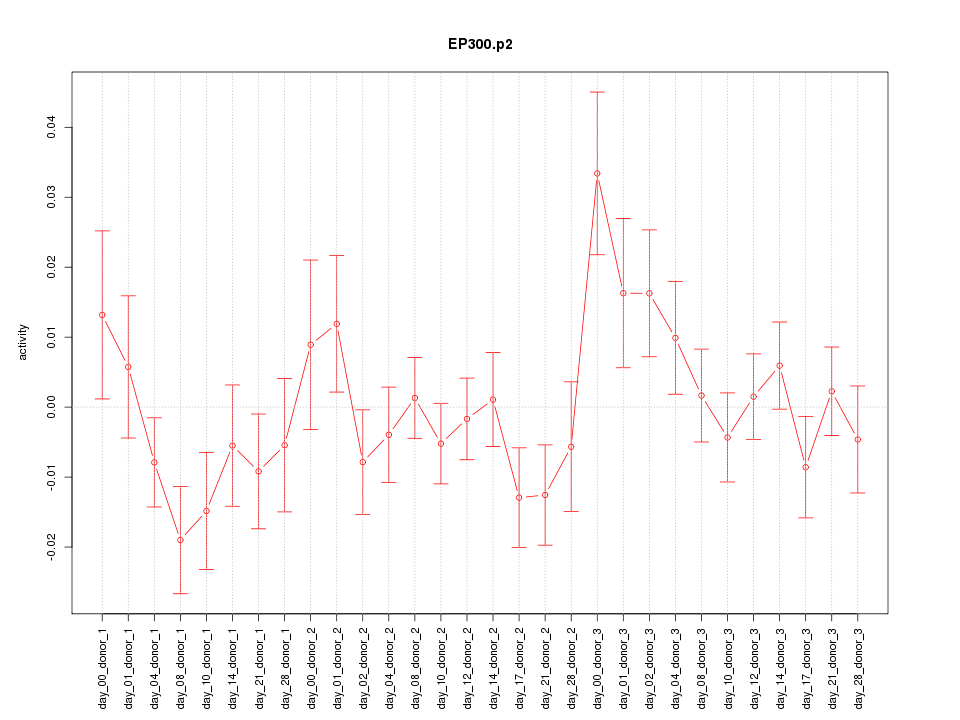
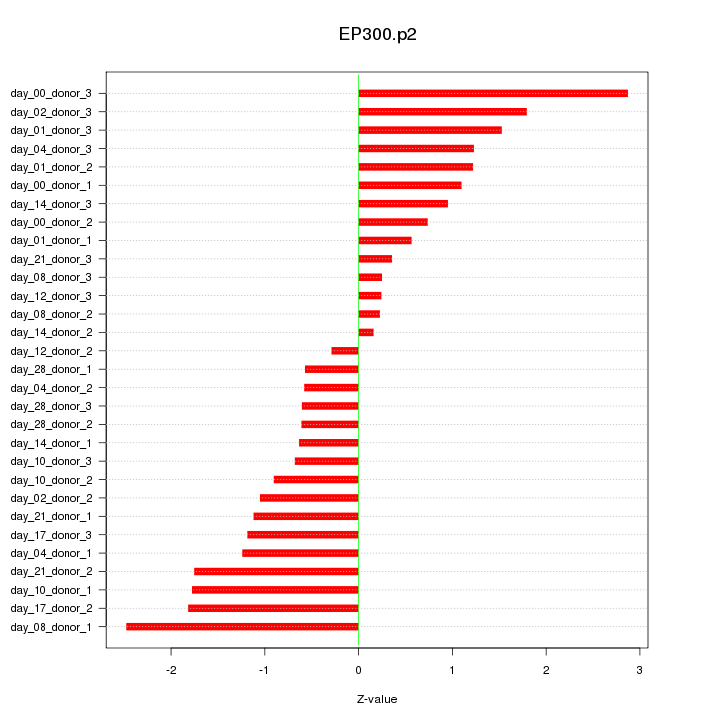
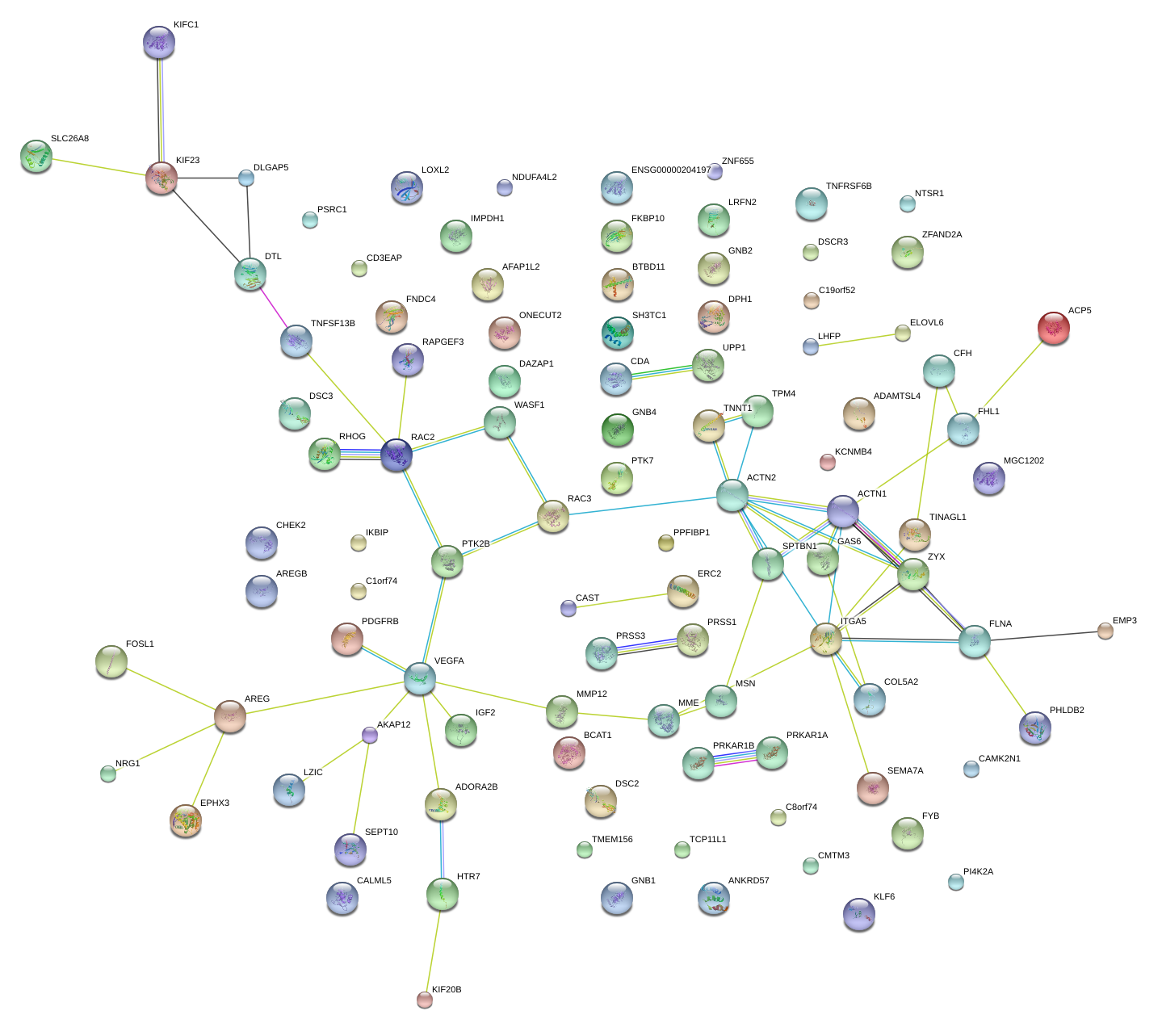
|
chr18_-_28681429
|
4.096
|
|
DSC2
|
desmocollin 2
|
|
chr18_-_28681885
|
3.354
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr12_-_25101993
|
2.763
|
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_54813002
|
2.581
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr9_+_33750463
|
2.155
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr19_-_11689591
|
2.041
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr12_-_54812936
|
1.998
|
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr22_-_37640187
|
1.895
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr2_+_110371887
|
1.887
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr1_+_20915440
|
1.871
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr1_-_109825683
|
1.823
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr7_-_752083
|
1.817
|
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr10_-_116164514
|
1.790
|
NM_001001936
NM_032550
|
AFAP1L2
|
actin filament associated protein 1-like 2
|
|
chr12_+_70759974
|
1.752
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr22_-_37640287
|
1.750
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr12_-_57634474
|
1.733
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chrX_+_135229536
|
1.665
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_+_48128939
|
1.621
|
|
UPP1
|
uridine phosphorylase 1
|
|
chr19_-_11689765
|
1.576
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr10_-_3827205
|
1.568
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr10_+_99400441
|
1.541
|
NM_018425
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr3_+_111578640
|
1.506
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr4_+_75480628
|
1.467
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr6_-_35992269
|
1.413
|
NM_138718
NM_001193476
NM_052961
|
SLC26A8
|
solute carrier family 26, member 8
|
|
chr4_+_75310852
|
1.410
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr7_-_128049959
|
1.389
|
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr20_+_62328002
|
1.388
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr1_-_20812727
|
1.384
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr8_-_23261626
|
1.380
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr6_+_43738757
|
1.366
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr17_+_15848230
|
1.354
|
NM_000676
|
ADORA2B
|
adenosine A2b receptor
|
|
chrX_+_135251454
|
1.329
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_-_209957872
|
1.323
|
NM_152485
|
C1orf74
|
chromosome 1 open reading frame 74
|
|
chr12_-_48152180
|
1.319
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr15_-_74726258
|
1.318
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr1_+_32043745
|
1.315
|
NM_001204415
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr8_-_23261632
|
1.311
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr2_+_54683276
|
1.276
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr3_+_154797435
|
1.265
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr10_-_3827418
|
1.264
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr4_+_8200918
|
1.254
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr7_-_128050006
|
1.246
|
NM_000883
NM_001102605
NM_001142576
NM_183243
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr8_-_23261591
|
1.226
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr7_+_99156263
|
1.212
|
|
ZNF655
|
zinc finger protein 655
|
|
chr11_-_2160199
|
1.212
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chrX_-_153599581
|
1.206
|
|
FLNA
|
filamin A, alpha
|
|
chr5_+_95998678
|
1.202
|
|
CAST
|
calpastatin
|
|
chr13_-_40177355
|
1.194
|
NM_005780
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr15_+_69706619
|
1.190
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chr14_-_69446038
|
1.180
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr17_+_39969205
|
1.173
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr14_-_69446008
|
1.167
|
|
ACTN1
|
actinin, alpha 1
|
|
chr21_-_38639593
|
1.165
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr3_-_179169180
|
1.161
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr8_+_31497267
|
1.157
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr13_-_114567040
|
1.150
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr6_-_40555111
|
1.142
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr6_+_33359312
|
1.140
|
NM_002263
|
KIFC1
|
kinesin family member C1
|
|
chrX_-_153599719
|
1.136
|
|
FLNA
|
filamin A, alpha
|
|
chr11_-_65667858
|
1.135
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr17_+_39969200
|
1.131
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr13_-_114567011
|
1.118
|
|
GAS6
|
growth arrest-specific 6
|
|
chr22_-_29137776
|
1.113
|
NM_001005735
NM_007194
NM_145862
|
CHEK2
|
checkpoint kinase 2
|
|
chr1_-_10003321
|
1.113
|
|
LZIC
|
leucine zipper and CTNNBIP1 domain containing
|
|
chr19_+_11039419
|
1.104
|
NM_138358
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr13_-_114566951
|
1.104
|
|
GAS6
|
growth arrest-specific 6
|
|
chr18_+_55102777
|
1.098
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr7_-_128045995
|
1.094
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr3_+_111578136
|
1.094
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr10_-_92617417
|
1.092
|
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr13_-_40177200
|
1.089
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr6_+_43043944
|
1.086
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr14_-_69445978
|
1.081
|
|
ACTN1
|
actinin, alpha 1
|
|
chr5_-_149535309
|
1.071
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr19_+_48828820
|
1.069
|
|
EMP3
|
epithelial membrane protein 3
|
|
chr19_-_15343767
|
1.060
|
NM_001142886
|
EPHX3
|
epoxide hydrolase 3
|
|
chr7_-_1199822
|
1.056
|
NM_182491
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr10_+_91461333
|
1.053
|
NM_016195
|
KIF20B
|
kinesin family member 20B
|
|
chr14_-_69445772
|
1.050
|
|
ACTN1
|
actinin, alpha 1
|
|
chr2_-_190044330
|
1.044
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr2_+_113816684
|
1.044
|
NM_012275
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr14_-_69445948
|
1.038
|
|
ACTN1
|
actinin, alpha 1
|
|
chr14_-_69445974
|
1.034
|
|
ACTN1
|
actinin, alpha 1
|
|
chr6_-_110501164
|
1.032
|
NM_001024934
NM_001024935
NM_001024936
NM_003931
|
WASF1
|
WAS protein family, member 1
|
|
chr5_-_39219578
|
1.026
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr14_-_69445989
|
1.019
|
|
ACTN1
|
actinin, alpha 1
|
|
chr8_+_27168953
|
1.017
|
NM_173174
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr4_-_111119752
|
1.014
|
NM_001130721
NM_024090
|
ELOVL6
|
ELOVL fatty acid elongase 6
|
|
chr4_-_39033981
|
1.011
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr12_-_99038705
|
1.008
|
|
IKBIP
|
IKBKB interacting protein
|
|
chr19_-_55658310
|
1.003
|
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr12_+_107712151
|
1.002
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr20_+_62328068
|
1.000
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr14_-_103995336
|
0.998
|
|
|
|
|
chr14_-_55658205
|
0.989
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr11_+_33061560
|
0.985
|
NM_001145541
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr13_-_114567034
|
0.981
|
|
GAS6
|
growth arrest-specific 6
|
|
chr7_-_1199785
|
0.980
|
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr2_-_27718098
|
0.977
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr20_+_61340165
|
0.971
|
NM_002531
|
NTSR1
|
neurotensin receptor 1 (high affinity)
|
|
chr3_+_154797644
|
0.970
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr2_-_110371404
|
0.969
|
|
SEPT10
|
septin 10
|
|
chr8_+_10530146
|
0.969
|
NM_001040032
|
C8orf74
|
chromosome 8 open reading frame 74
|
|
chr1_+_212209129
|
0.967
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr2_-_27718059
|
0.959
|
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr16_-_86542463
|
0.952
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr8_-_23261675
|
0.943
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr6_+_151561653
|
0.938
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chrX_+_64887510
|
0.937
|
NM_002444
|
MSN
|
moesin
|
|
chr17_+_1933365
|
0.933
|
NM_001383
|
DPH1
|
DPH1 homolog (S. cerevisiae)
|
|
chr16_+_66638652
|
0.931
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr10_-_5541401
|
0.929
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr7_+_143078395
|
0.926
|
|
ZYX
|
zyxin
|
|
chr2_-_110371669
|
0.926
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr1_+_150521786
|
0.912
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr7_+_143078358
|
0.906
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr13_-_40177304
|
0.903
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr19_+_16187315
|
0.898
|
|
TPM4
|
tropomyosin 4
|
|
chr12_+_27677044
|
0.897
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr12_-_54813010
|
0.896
|
NM_002205
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide)
|
|
chr11_-_3862183
|
0.894
|
NM_001665
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr7_-_1199813
|
0.890
|
|
ZFAND2A
|
zinc finger, AN1-type domain 2A
|
|
chr10_+_99400469
|
0.887
|
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr11_+_33060962
|
0.884
|
NM_018393
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr1_-_154946830
|
0.884
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr20_-_56286540
|
0.882
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr3_+_156392204
|
0.875
|
NM_015508
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr20_-_10654429
|
0.875
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr1_-_175161832
|
0.875
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr3_+_111578026
|
0.874
|
NM_001134438
NM_145753
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr12_-_48152860
|
0.873
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr8_+_32504250
|
0.872
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr17_+_76210276
|
0.872
|
NM_001012270
NM_001012271
NM_001168
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr3_-_13921617
|
0.868
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chr3_-_51996855
|
0.866
|
NM_033010
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr19_+_45843997
|
0.862
|
NM_177417
|
KLC3
|
kinesin light chain 3
|
|
chr15_-_81616439
|
0.859
|
NM_181900
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5
|
|
chr3_+_19189969
|
0.858
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr12_-_49365545
|
0.856
|
NM_003394
|
WNT10B
|
wingless-type MMTV integration site family, member 10B
|
|
chr5_+_31193761
|
0.852
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr22_+_31063914
|
0.848
|
|
|
|
|
chr15_+_40453263
|
0.848
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr11_+_706607
|
0.847
|
|
EPS8L2
|
EPS8-like 2
|
|
chr3_+_156392714
|
0.847
|
NM_001184717
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr11_-_65667860
|
0.847
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr11_+_35160385
|
0.845
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
NM_001202555
NM_001202556
NM_001202557
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr1_-_113498828
|
0.844
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr17_-_4851797
|
0.844
|
NM_005022
|
PFN1
|
profilin 1
|
|
chrX_-_54522318
|
0.843
|
NM_004463
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr7_-_752132
|
0.835
|
NM_002735
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta
|
|
chr8_-_145013628
|
0.833
|
NM_201384
|
PLEC
|
plectin
|
|
chr6_-_42162693
|
0.833
|
NM_002098
|
GUCA1B
|
guanylate cyclase activator 1B (retina)
|
|
chr2_+_113816214
|
0.832
|
NM_173170
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr11_-_2925174
|
0.830
|
NM_007105
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense
|
|
chr19_-_41859253
|
0.829
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr3_+_136537860
|
0.829
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr1_-_155224668
|
0.829
|
|
FAM189B
|
family with sequence similarity 189, member B
|
|
chr6_+_46655611
|
0.827
|
NM_001010870
NM_001168359
|
TDRD6
|
tudor domain containing 6
|
|
chr22_-_37915209
|
0.827
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr9_-_34590137
|
0.825
|
NM_001207011
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr11_-_46722139
|
0.825
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr6_+_43738394
|
0.823
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr11_+_33061291
|
0.817
|
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr2_+_191745912
|
0.815
|
|
GLS
|
glutaminase
|
|
chr1_-_154946859
|
0.814
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr22_-_39095991
|
0.812
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr11_+_129245834
|
0.811
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr1_-_109825745
|
0.811
|
|
|
|
|
chr1_+_26606621
|
0.804
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr15_+_67358194
|
0.803
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr1_+_153651050
|
0.801
|
NM_000906
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A)
|
|
chr1_-_95007139
|
0.800
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr11_-_102962600
|
0.793
|
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae)
|
|
chr1_+_1550794
|
0.792
|
NM_001170686
NM_001170687
NM_001170688
NM_080875
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr7_+_116165089
|
0.789
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr2_-_9695827
|
0.788
|
|
ADAM17
|
ADAM metallopeptidase domain 17
|
|
chr8_+_22022644
|
0.785
|
NM_001199
NM_006129
|
BMP1
|
bone morphogenetic protein 1
|
|
chr1_+_222886655
|
0.785
|
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr12_+_6937764
|
0.784
|
|
LEPREL2
|
leprecan-like 2
|
|
chr1_-_197115566
|
0.778
|
NM_001206846
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr2_+_31456879
|
0.777
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr16_-_10275743
|
0.773
|
NM_001134408
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr17_-_43209681
|
0.770
|
NM_133373
|
PLCD3
|
phospholipase C, delta 3
|
|
chr1_-_154946801
|
0.761
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr9_-_129884961
|
0.761
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr1_+_206138909
|
0.760
|
NM_001123168
|
FAM72B
FAM72A
|
family with sequence similarity 72, member B
family with sequence similarity 72, member A
|
|
chrX_-_100662788
|
0.755
|
|
GLA
|
galactosidase, alpha
|
|
chr11_+_61595633
|
0.754
|
NM_004265
|
FADS2
|
fatty acid desaturase 2
|
|
chr8_-_23021447
|
0.754
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr5_-_60140041
|
0.752
|
NM_001104558
NM_024930
|
ELOVL7
|
ELOVL fatty acid elongase 7
|
|
chr1_+_212208918
|
0.752
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr15_+_40453209
|
0.750
|
NM_001211
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr4_-_40859081
|
0.750
|
NM_001166051
NM_001166052
NM_001166053
NM_001166054
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr14_+_53019865
|
0.749
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr1_+_26606580
|
0.747
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr20_+_44034632
|
0.746
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr7_+_143078457
|
0.743
|
|
ZYX
|
zyxin
|
|
chr15_-_75871508
|
0.743
|
NM_002833
|
PTPN9
|
protein tyrosine phosphatase, non-receptor type 9
|
|
chr19_-_19052032
|
0.743
|
NM_004838
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chrX_-_154033729
|
0.743
|
NM_001166460
NM_001166461
NM_001166462
NM_002436
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa
|