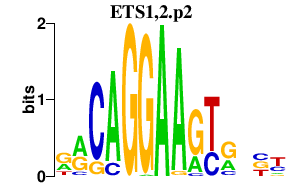
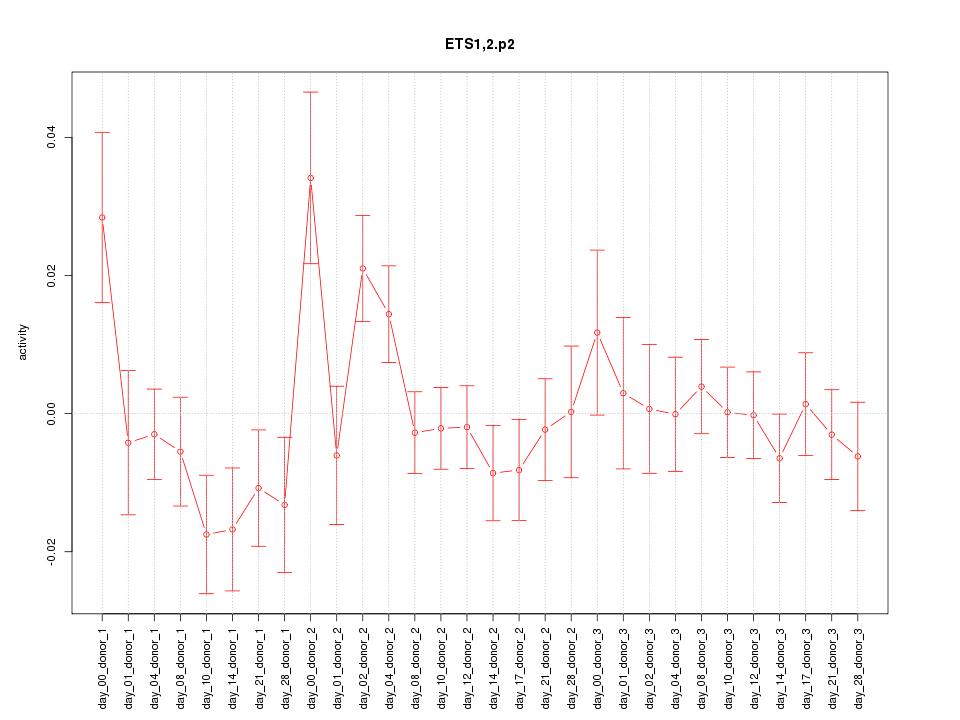
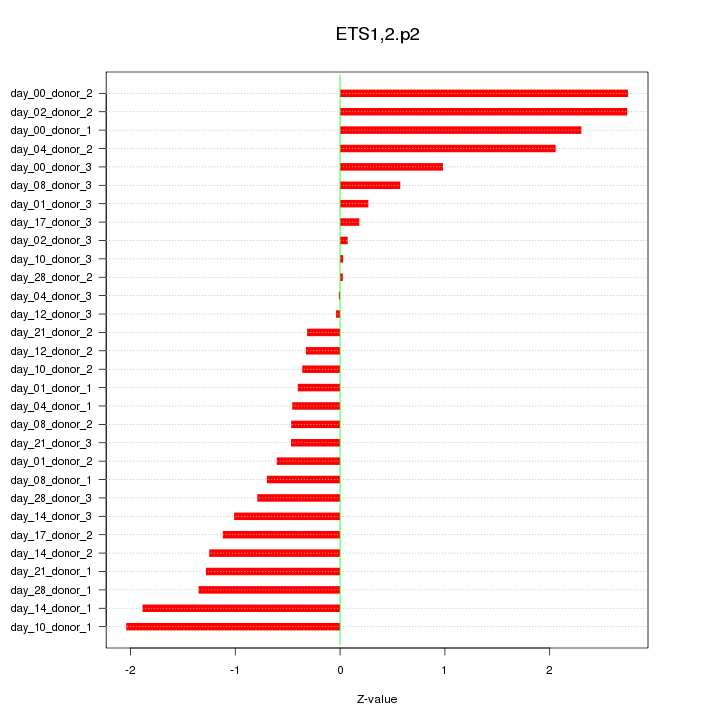
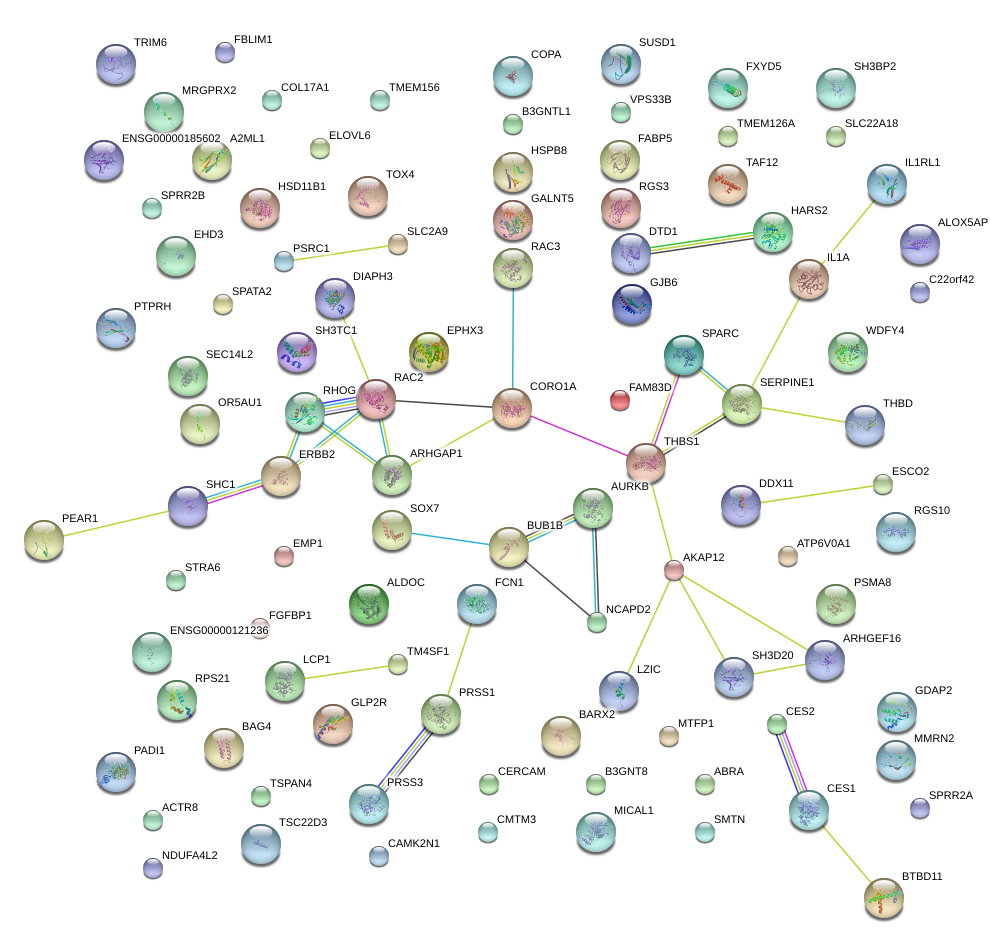
|
chr1_-_154946859
|
3.828
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr13_-_20805371
|
3.289
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr1_-_154946830
|
3.212
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr12_-_57634474
|
3.014
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr5_-_151066445
|
2.807
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr10_-_88717319
|
2.772
|
NM_024756
|
MMRN2
|
multimerin 2
|
|
chr1_-_154946801
|
2.762
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr4_+_2813966
|
2.684
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr1_-_153044083
|
2.615
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr17_-_8113862
|
2.347
|
NM_004217
|
AURKB
|
aurora kinase B
|
|
chr19_+_35646006
|
2.307
|
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr2_+_102953716
|
2.302
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr19_+_35645640
|
2.301
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr19_+_35645614
|
2.270
|
NM_014164
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr5_-_151066492
|
2.172
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr11_+_2920910
|
2.151
|
NM_183233
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr13_-_60738078
|
2.117
|
NM_001042517
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr17_+_9729380
|
2.097
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr12_+_8975149
|
1.991
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr1_+_16083132
|
1.988
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr10_-_105845619
|
1.898
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr10_-_121302149
|
1.846
|
NM_001005339
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr22_+_30792929
|
1.824
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr1_-_153029987
|
1.777
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr4_+_2813945
|
1.762
|
NM_001145855
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr15_-_74500394
|
1.735
|
NM_001199041
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr2_+_31456879
|
1.717
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr19_-_15343194
|
1.714
|
NM_024794
|
EPHX3
|
epoxide hydrolase 3
|
|
chr20_+_60962120
|
1.706
|
NM_001024
|
RPS21
|
ribosomal protein S21
|
|
chr9_+_131185129
|
1.700
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr1_+_156863522
|
1.681
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr22_-_37640187
|
1.648
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr6_+_151561094
|
1.639
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr20_-_48532036
|
1.620
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr10_-_105845556
|
1.588
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr2_-_113542970
|
1.577
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr4_-_15939949
|
1.569
|
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr2_+_158114109
|
1.565
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr1_-_118472178
|
1.560
|
|
GDAP2
|
ganglioside induced differentiation associated protein 2
|
|
chr7_+_100770378
|
1.533
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr22_-_37640287
|
1.514
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr9_+_116356407
|
1.499
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr4_+_8200918
|
1.466
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr1_+_17531619
|
1.445
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr1_+_209859524
|
1.440
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr17_+_40610940
|
1.430
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr22_+_31480981
|
1.417
|
NM_001207018
|
SMTN
|
smoothelin
|
|
chr12_+_13349720
|
1.399
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr22_-_32555200
|
1.377
|
NM_001010859
|
C22orf42
|
chromosome 22 open reading frame 42
|
|
chr15_-_91565685
|
1.373
|
NM_018668
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast)
|
|
chr9_-_137809710
|
1.371
|
NM_002003
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1
|
|
chr13_+_31287614
|
1.368
|
NM_001204406
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein
|
|
chr4_-_111119752
|
1.366
|
NM_001130721
NM_024090
|
ELOVL6
|
ELOVL fatty acid elongase 6
|
|
chr8_-_10587942
|
1.361
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr11_-_46722139
|
1.359
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr4_-_10023094
|
1.356
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr12_-_57630867
|
1.355
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr1_-_10003321
|
1.349
|
|
LZIC
|
leucine zipper and CTNNBIP1 domain containing
|
|
chr8_+_82192717
|
1.347
|
NM_001444
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated)
|
|
chr6_-_109776946
|
1.343
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr6_-_109776942
|
1.335
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr17_+_40610897
|
1.334
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr9_+_116356199
|
1.328
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr7_+_142457318
|
1.323
|
NM_002769
|
PRSS1
PRSS2
|
protease, serine, 1 (trypsin 1)
protease, serine, 2 (trypsin 2)
|
|
chr19_-_55720812
|
1.320
|
NM_001161440
NM_002842
|
PTPRH
|
protein tyrosine phosphatase, receptor type, H
|
|
chr11_+_129245834
|
1.311
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr16_+_66638530
|
1.311
|
NM_181553
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr9_-_114937464
|
1.310
|
NM_022486
|
SUSD1
|
sushi domain containing 1
|
|
chr17_+_40610875
|
1.294
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr20_-_23030141
|
1.292
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr15_+_39873276
|
1.279
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr16_+_66968390
|
1.279
|
|
CES2
|
carboxylesterase 2
|
|
chr20_+_37555055
|
1.272
|
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr12_+_107712151
|
1.270
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr12_+_13349601
|
1.267
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr4_-_15940362
|
1.262
|
NM_005130
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr20_+_37554954
|
1.255
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr4_-_39033981
|
1.248
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr17_-_26903854
|
1.244
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr12_+_6602521
|
1.197
|
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr17_-_43510204
|
1.196
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr1_+_3388170
|
1.195
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr11_+_5617330
|
1.192
|
NM_058166
|
TRIM6
|
tripartite motif containing 6
|
|
chr5_+_140071220
|
1.174
|
|
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr12_+_119616594
|
1.169
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr13_-_46756291
|
1.163
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr16_+_30194925
|
1.160
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr15_+_40453263
|
1.142
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr19_-_41934634
|
1.142
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr1_-_28969516
|
1.142
|
NM_001135218
NM_005644
|
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa
|
|
chr3_-_149095458
|
1.130
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr15_+_40453209
|
1.127
|
NM_001211
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chrX_-_107019001
|
1.114
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr16_+_66638652
|
1.110
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr6_-_109777171
|
1.108
|
NM_001159291
NM_022765
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr1_-_20812727
|
1.106
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr11_+_844445
|
1.104
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr18_+_23713815
|
1.102
|
NM_001025096
NM_001025097
NM_144662
|
PSMA8
|
proteasome (prosome, macropain) subunit, alpha type, 8
|
|
chr11_-_46722115
|
1.099
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr1_-_109825767
|
1.098
|
NM_001005290
NM_001032291
NM_032636
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr1_-_109825683
|
1.093
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr10_+_49892860
|
1.089
|
|
WDFY4
|
WDFY family member 4
|
|
chr11_-_19082227
|
1.088
|
NM_054030
|
MRGPRX2
|
MAS-related GPR, member X2
|
|
chr1_-_160313056
|
1.084
|
|
COPA
|
coatomer protein complex, subunit alpha
|
|
chr17_+_40610905
|
1.084
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr13_-_60737899
|
1.083
|
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr8_+_38034066
|
1.080
|
NM_001204878
NM_004874
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr3_-_139258373
|
1.076
|
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr16_+_1756161
|
1.065
|
NM_001040439
NM_015133
|
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr2_+_37571752
|
1.063
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr15_-_80263460
|
1.058
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr17_+_40610915
|
1.056
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr3_-_149095413
|
1.056
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr15_-_40600065
|
1.053
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr11_+_46722564
|
1.053
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr6_-_30654978
|
1.052
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr3_+_47324329
|
1.052
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr7_+_18548899
|
1.051
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr19_+_58694444
|
1.034
|
|
ZNF274
|
zinc finger protein 274
|
|
chr5_+_140071014
|
1.034
|
NM_012208
|
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr3_-_149095517
|
1.033
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr5_+_140071141
|
1.030
|
|
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr16_+_66968319
|
1.020
|
NM_003869
NM_198061
|
CES2
|
carboxylesterase 2
|
|
chr21_-_40685545
|
1.013
|
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr17_+_40610899
|
1.012
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr17_-_39093671
|
1.011
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr9_-_88969256
|
1.010
|
|
ZCCHC6
|
zinc finger, CCHC domain containing 6
|
|
chr5_+_96212167
|
0.998
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr3_-_139258477
|
0.990
|
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr2_-_208031570
|
0.988
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr19_+_10397642
|
0.982
|
NM_001039132
NM_001544
NM_022377
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)
|
|
chr16_+_31483066
|
0.981
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr22_+_33196801
|
0.979
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr22_+_38864015
|
0.974
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr7_+_2281856
|
0.973
|
NM_002452
NM_198948
NM_198949
NM_198950
NM_198952
|
NUDT1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1
|
|
chr19_+_17862285
|
0.966
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr1_-_6420719
|
0.964
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr9_+_34652125
|
0.960
|
NM_001142784
|
IL11RA
|
interleukin 11 receptor, alpha
|
|
chr2_+_234160216
|
0.959
|
NM_001190266
NM_001190267
NM_017974
NM_030803
NM_198890
|
ATG16L1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae)
|
|
chr16_+_66968360
|
0.959
|
|
CES2
|
carboxylesterase 2
|
|
chr6_-_41130820
|
0.952
|
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr1_-_162381805
|
0.951
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr11_-_2170792
|
0.947
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr15_+_91473409
|
0.945
|
NM_001039675
|
UNC45A
|
unc-45 homolog A (C. elegans)
|
|
chr16_+_29465821
|
0.943
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 readthrough
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr15_+_93447702
|
0.942
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr6_-_42162693
|
0.933
|
NM_002098
|
GUCA1B
|
guanylate cyclase activator 1B (retina)
|
|
chr18_+_61420276
|
0.933
|
NM_001040147
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr2_+_234160345
|
0.932
|
|
ATG16L1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae)
|
|
chr14_-_67878859
|
0.932
|
|
PLEK2
|
pleckstrin 2
|
|
chr6_-_41130864
|
0.928
|
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr1_-_20987937
|
0.926
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr2_-_177502658
|
0.921
|
|
LOC375295
|
uncharacterized LOC375295
|
|
chr1_+_1550794
|
0.920
|
NM_001170686
NM_001170687
NM_001170688
NM_080875
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr1_-_28520379
|
0.916
|
NM_001164721
NM_001164722
|
PTAFR
|
platelet-activating factor receptor
|
|
chr8_+_22462267
|
0.914
|
|
KIAA1967
|
KIAA1967
|
|
chr9_+_77703412
|
0.913
|
|
OSTF1
|
osteoclast stimulating factor 1
|
|
chr14_-_103995336
|
0.912
|
|
|
|
|
chr17_+_40610857
|
0.910
|
NM_001130020
NM_001130021
NM_005177
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr20_-_42839295
|
0.909
|
|
C20orf111
|
chromosome 20 open reading frame 111
|
|
chr11_+_124933007
|
0.908
|
NM_001145290
NM_198277
|
SLC37A2
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 2
|
|
chr14_-_75422466
|
0.904
|
NM_001207012
NM_002632
|
PGF
|
placental growth factor
|
|
chr15_-_40599974
|
0.903
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_+_6086369
|
0.900
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr18_+_47088400
|
0.899
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr1_-_212588239
|
0.898
|
NM_001198862
NM_018252
|
TMEM206
|
transmembrane protein 206
|
|
chr10_+_17271302
|
0.896
|
|
VIM
|
vimentin
|
|
chr17_-_5138049
|
0.891
|
NM_207103
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chr11_+_46722316
|
0.889
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr13_+_46039024
|
0.888
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr7_-_107643330
|
0.885
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr19_+_58694381
|
0.877
|
NM_016324
NM_016325
NM_133502
|
ZNF274
|
zinc finger protein 274
|
|
chr1_-_160312959
|
0.876
|
|
|
|
|
chr1_-_6580068
|
0.876
|
NM_198681
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr12_+_1800146
|
0.875
|
|
ADIPOR2
|
adiponectin receptor 2
|
|
chr9_-_35103089
|
0.873
|
|
STOML2
|
stomatin (EPB72)-like 2
|
|
chr19_+_45281125
|
0.872
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr14_-_67878770
|
0.872
|
NM_016445
|
PLEK2
|
pleckstrin 2
|
|
chr2_-_110371404
|
0.871
|
|
SEPT10
|
septin 10
|
|
chr1_-_6545521
|
0.867
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr12_+_107974633
|
0.864
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr2_+_233925104
|
0.859
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr16_+_66968365
|
0.853
|
|
CES2
|
carboxylesterase 2
|
|
chr9_+_125133228
|
0.851
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr7_+_80267782
|
0.844
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr14_-_100842637
|
0.841
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr8_+_74903563
|
0.839
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr12_+_53440809
|
0.832
|
NM_198316
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr1_-_24126031
|
0.831
|
NM_001127621
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr1_+_6086412
|
0.827
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr19_+_41257086
|
0.825
|
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr2_-_110371669
|
0.820
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr11_+_66824285
|
0.817
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr8_+_38034296
|
0.815
|
|
BAG4
|
BCL2-associated athanogene 4
|
|
chr6_+_31515352
|
0.814
|
NM_001144961
NM_005007
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1
|
|
chrX_-_40594663
|
0.811
|
|
MED14
|
mediator complex subunit 14
|
|
chr4_+_153701113
|
0.810
|
|
ARFIP1
|
ADP-ribosylation factor interacting protein 1
|
|
chr10_-_98031154
|
0.810
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr6_-_41130921
|
0.807
|
NM_018965
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr2_+_233925035
|
0.806
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|