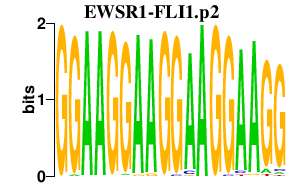
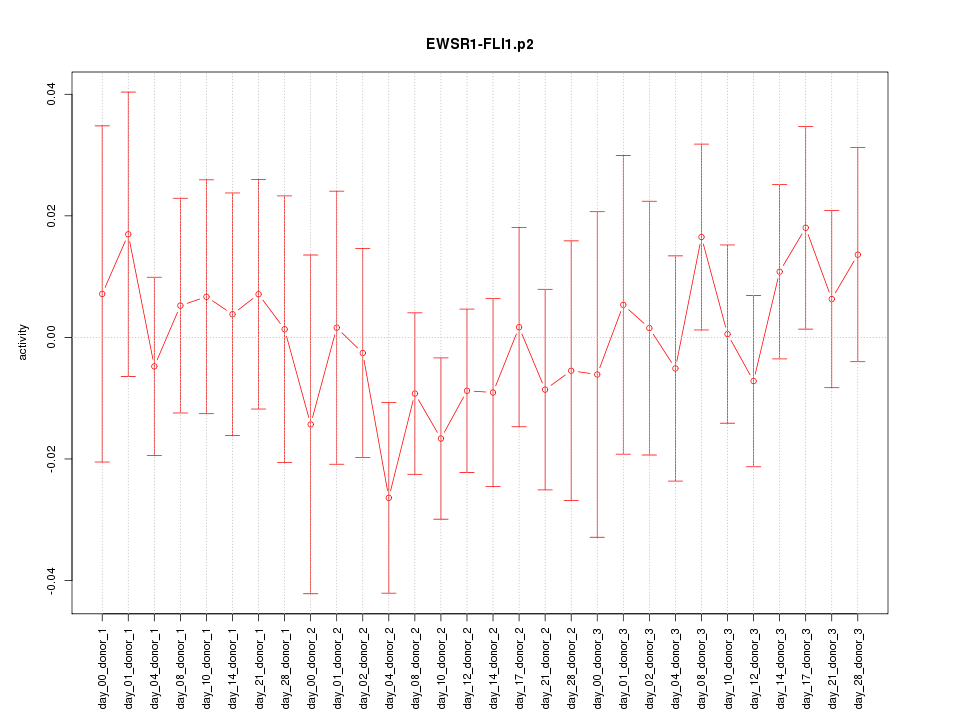
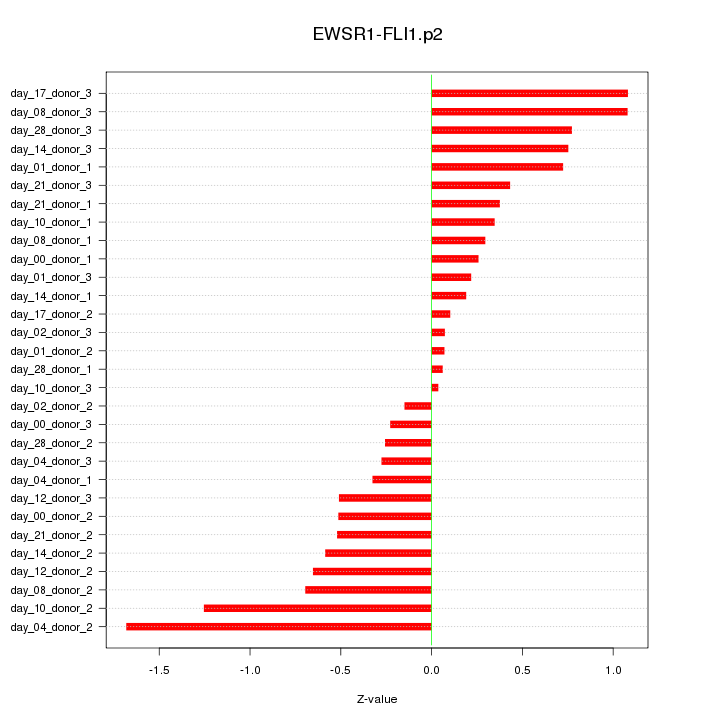
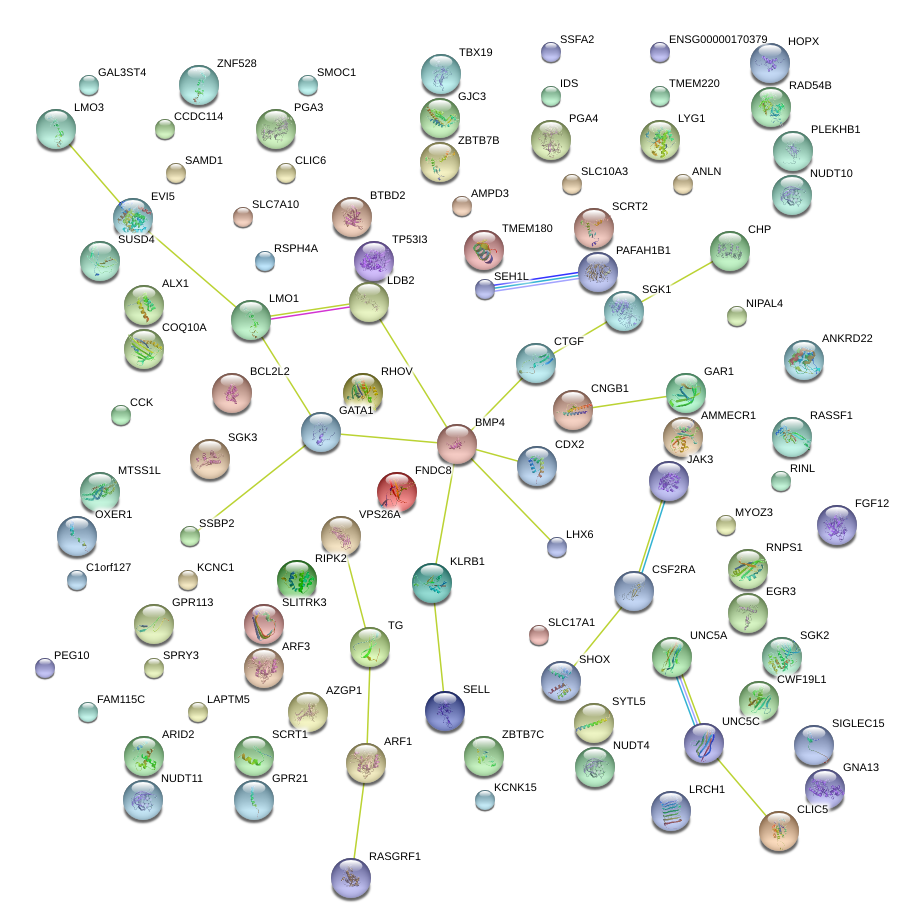
|
chrX_+_37892786
|
1.658
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr3_-_42307661
|
0.987
|
NM_000729
|
CCK
|
cholecystokinin
|
|
chr18_+_43405544
|
0.737
|
NM_213602
|
SIGLEC15
|
sialic acid binding Ig-like lectin 15
|
|
chrX_+_585078
|
0.644
|
NM_000451
NM_006883
|
SHOX
|
short stature homeobox
|
|
chrY_+_535078
|
0.636
|
NM_000451
NM_006883
|
SHOX
|
short stature homeobox
|
|
chr9_-_124990706
|
0.616
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr6_-_25832268
|
0.570
|
NM_005074
|
SLC17A1
|
solute carrier family 17 (sodium phosphate), member 1
|
|
chrX_-_109561427
|
0.534
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr20_+_43374487
|
0.514
|
NM_022358
|
KCNK15
|
potassium channel, subfamily K, member 15
|
|
chr6_+_116937641
|
0.510
|
NM_001010892
NM_001161664
|
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas)
|
|
chr14_-_54423448
|
0.505
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr3_-_164914448
|
0.469
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr2_-_99917638
|
0.467
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr20_+_42187634
|
0.457
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr8_+_133879202
|
0.456
|
NM_003235
|
TG
|
thyroglobulin
|
|
chr7_-_99527242
|
0.420
|
NM_181538
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa
|
|
chrX_-_109561298
|
0.403
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr5_+_156887026
|
0.378
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chrX_+_1401570
|
0.371
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr7_+_36429388
|
0.368
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chrY_+_1351570
|
0.355
|
NM_001161530
NM_172247
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr6_-_132272311
|
0.344
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr17_-_10633645
|
0.342
|
NM_001004313
|
TMEM220
|
transmembrane protein 220
|
|
chr1_-_169680677
|
0.329
|
NM_000655
|
SELL
|
selectin L
|
|
chr9_-_124989755
|
0.326
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr7_-_99573637
|
0.323
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr11_+_73357222
|
0.313
|
NM_001130033
NM_021200
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr6_-_134497069
|
0.312
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr3_-_192445344
|
0.308
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr6_-_134496833
|
0.303
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr15_-_79298001
|
0.284
|
NM_153815
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr12_+_85673884
|
0.280
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr19_-_33716609
|
0.275
|
NM_019849
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10
|
|
chrX_-_109561314
|
0.273
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr12_+_93771607
|
0.262
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr2_-_105467912
|
0.261
|
|
LOC100506421
|
uncharacterized LOC100506421
|
|
chr12_-_9760481
|
0.261
|
NM_002258
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1
|
|
chr19_-_48823331
|
0.258
|
NM_144577
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr3_-_50378238
|
0.258
|
NM_001206957
NM_007182
NM_170714
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr2_-_24307529
|
0.252
|
NM_004881
NM_147184
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr18_+_12948004
|
0.250
|
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr11_+_60970983
|
0.249
|
NM_001079807
|
PGA3
PGA4
|
pepsinogen 3, group I (pepsinogen A)
pepsinogen 4, group I (pepsinogen A)
|
|
chr8_-_145559831
|
0.245
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr10_-_90611675
|
0.241
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr1_-_93257950
|
0.234
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr5_-_81046943
|
0.234
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr17_+_2497233
|
0.233
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr12_+_56660913
|
0.231
|
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr3_-_192126837
|
0.229
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr12_+_56661026
|
0.224
|
NM_001099337
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr12_+_56660548
|
0.204
|
NM_144576
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr4_-_16900348
|
0.202
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr12_+_56660724
|
0.201
|
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr9_+_125796845
|
0.196
|
NM_005294
|
GPR21
|
G protein-coupled receptor 21
|
|
chr4_-_16900028
|
0.195
|
|
LDB2
|
LIM domain binding 2
|
|
chr19_+_52901101
|
0.193
|
NM_032423
|
ZNF528
|
zinc finger protein 528
|
|
chr17_+_33448597
|
0.193
|
NM_017559
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr11_+_10476665
|
0.192
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr8_-_95487284
|
0.189
|
NM_001205262
NM_012415
|
RAD54B
|
RAD54 homolog B (S. cerevisiae)
|
|
chr11_+_10477480
|
0.188
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr16_-_70719854
|
0.182
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr7_-_99766191
|
0.181
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr2_+_113816214
|
0.180
|
NM_173170
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chrX_-_148586666
|
0.178
|
NM_000202
NM_001166550
NM_006123
|
IDS
|
iduronate 2-sulfatase
|
|
chr19_-_14201230
|
0.177
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr2_-_26541916
|
0.175
|
NM_001145168
NM_001145169
|
GPR113
|
G protein-coupled receptor 113
|
|
chr14_+_70346113
|
0.173
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr2_+_113816684
|
0.173
|
NM_012275
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr7_+_143412938
|
0.173
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr4_-_57522469
|
0.167
|
NM_001145459
NM_139211
|
HOPX
|
HOP homeobox
|
|
chr18_+_12947972
|
0.162
|
NM_001013437
NM_031216
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr1_-_223537400
|
0.160
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr2_-_42991201
|
0.156
|
NM_148962
|
OXER1
|
oxoeicosanoid (OXE) receptor 1
|
|
chr5_+_150040402
|
0.155
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr19_-_17958814
|
0.155
|
NM_000215
|
JAK3
|
Janus kinase 3
|
|
chr12_-_16759939
|
0.154
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr16_-_2318412
|
0.152
|
NM_080594
|
RNPS1
|
RNA binding protein S1, serine-rich domain
|
|
chr10_+_104221158
|
0.151
|
NM_024789
|
TMEM180
|
transmembrane protein 180
|
|
chr1_-_11042093
|
0.151
|
NM_001170754
|
C1orf127
|
chromosome 1 open reading frame 127
|
|
chr1_+_154975201
|
0.143
|
NM_001252406
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr13_+_47127383
|
0.140
|
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1
|
|
chr13_-_28543290
|
0.138
|
NM_001265
|
CDX2
|
caudal type homeobox 2
|
|
chr7_-_99766238
|
0.136
|
NM_024637
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr11_+_17757494
|
0.136
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr6_-_45983279
|
0.136
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr18_-_45935627
|
0.136
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr1_-_31230594
|
0.135
|
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr12_+_46123495
|
0.132
|
NM_152641
|
ARID2
|
AT rich interactive domain 2 (ARID, RFX-like)
|
|
chr1_-_31230673
|
0.130
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chrX_+_154997396
|
0.130
|
NM_005840
|
SPRY3
|
sprouty homolog 3 (Drosophila)
|
|
chr4_-_96470114
|
0.127
|
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr19_-_2015628
|
0.124
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr19_-_39368889
|
0.124
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr18_+_12948010
|
0.123
|
|
SEH1L
|
SEH1-like (S. cerevisiae)
|
|
chr2_+_182756468
|
0.120
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chrX_+_48644961
|
0.118
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr17_-_63052816
|
0.118
|
|
|
|
|
chr4_-_16900256
|
0.117
|
|
LDB2
|
LIM domain binding 2
|
|
chr8_-_22550057
|
0.116
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chrX_-_153718984
|
0.115
|
NM_001142391
NM_001142392
NM_019848
|
SLC10A3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3
|
|
chr17_-_63052707
|
0.115
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr16_+_67022487
|
0.114
|
NM_173815
|
CES4A
|
carboxylesterase 4A
|
|
chr7_+_94285620
|
0.114
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr19_-_17958773
|
0.112
|
|
JAK3
|
Janus kinase 3
|
|
chr10_+_70883930
|
0.110
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr14_+_23775970
|
0.109
|
NM_001199864
NM_004050
NM_001199839
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough
BCL2-like 2
|
|
chr13_+_47127312
|
0.108
|
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1
|
|
chr12_-_49351240
|
0.108
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr10_-_102027337
|
0.107
|
NM_018294
|
CWF19L1
|
CWF19-like 1, cell cycle control (S. pombe)
|
|
chr4_+_110736636
|
0.107
|
NM_018983
NM_032993
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr5_-_81046858
|
0.107
|
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr15_+_41523465
|
0.107
|
|
CHP
|
calcium binding protein P22
|
|
chr8_-_37823986
|
0.106
|
NM_000025
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr18_+_46065367
|
0.105
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chrX_+_198060
|
0.103
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr13_+_47127295
|
0.102
|
NM_001164211
NM_001164213
NM_015116
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1
|
|
chr12_-_117175844
|
0.102
|
|
C12orf49
|
chromosome 12 open reading frame 49
|
|
chr11_-_67407024
|
0.101
|
NM_005995
|
TBX10
|
T-box 10
|
|
chr1_-_26232643
|
0.100
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr10_+_70883943
|
0.100
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chrY_+_148060
|
0.099
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr10_+_70883987
|
0.099
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr19_+_13205956
|
0.098
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr1_+_186798031
|
0.098
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr1_-_26232890
|
0.097
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr11_+_60989820
|
0.097
|
NM_001079808
|
PGA3
PGA4
|
pepsinogen 3, group I (pepsinogen A)
pepsinogen 4, group I (pepsinogen A)
|
|
chr2_+_149402321
|
0.096
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chrX_-_153718937
|
0.096
|
|
SLC10A3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3
|
|
chr12_-_49351263
|
0.096
|
|
|
|
|
chr10_+_70883977
|
0.093
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr8_-_27695543
|
0.092
|
|
PBK
|
PDZ binding kinase
|
|
chr17_+_635656
|
0.091
|
NM_024792
|
FAM57A
|
family with sequence similarity 57, member A
|
|
chr4_+_154125589
|
0.090
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_+_149402559
|
0.090
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr1_+_154975041
|
0.089
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr11_+_101980736
|
0.089
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr1_+_167599329
|
0.089
|
NM_052862
|
RCSD1
|
RCSD domain containing 1
|
|
chr1_+_44412418
|
0.088
|
NM_014652
|
IPO13
|
importin 13
|
|
chr12_-_49351129
|
0.088
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chrX_+_152599544
|
0.088
|
NM_001080485
|
ZNF275
|
zinc finger protein 275
|
|
chr8_-_145701717
|
0.087
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr17_-_63052884
|
0.086
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr12_+_49658865
|
0.086
|
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr2_+_182756665
|
0.084
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr4_-_85887543
|
0.083
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr10_+_70883856
|
0.082
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr22_+_19467413
|
0.081
|
NM_001178010
NM_001178011
NM_003504
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr6_+_52284948
|
0.080
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr5_-_142000899
|
0.079
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr3_+_170075494
|
0.079
|
|
SKIL
|
SKI-like oncogene
|
|
chr14_-_21566728
|
0.078
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr11_-_65488142
|
0.076
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr6_-_24719286
|
0.076
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr15_-_68498373
|
0.075
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr9_+_115513050
|
0.074
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr12_-_49351245
|
0.074
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr8_-_27695288
|
0.074
|
NM_018492
|
PBK
|
PDZ binding kinase
|
|
chr7_+_94285736
|
0.071
|
|
PEG10
|
paternally expressed 10
|
|
chr1_-_152009406
|
0.070
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr17_-_40833840
|
0.068
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr19_+_8117645
|
0.067
|
NM_001201359
NM_005624
|
CCL25
|
chemokine (C-C motif) ligand 25
|
|
chr10_-_71892596
|
0.067
|
NM_001198696
NM_032797
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2
|
|
chr16_-_74734687
|
0.067
|
NM_001142497
NM_152649
|
MLKL
|
mixed lineage kinase domain-like
|
|
chr1_+_246887591
|
0.065
|
|
SCCPDH
|
saccharopine dehydrogenase (putative)
|
|
chr17_+_48133357
|
0.065
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr1_-_154193044
|
0.065
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr9_-_130639939
|
0.065
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr12_-_49351297
|
0.064
|
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr1_-_152009440
|
0.064
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr12_+_49658862
|
0.064
|
NM_032704
|
TUBA1C
|
tubulin, alpha 1c
|
|
chr18_+_10454542
|
0.063
|
|
APCDD1
|
adenomatosis polyposis coli down-regulated 1
|
|
chr16_+_69458489
|
0.061
|
NM_030579
|
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane)
|
|
chr19_+_16144929
|
0.061
|
|
FLJ25328
|
uncharacterized LOC148231
|
|
chr11_-_123065836
|
0.061
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr19_+_1207129
|
0.060
|
|
EFNA2
|
ephrin-A2
|
|
chr11_+_71927818
|
0.060
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chr6_+_36916038
|
0.059
|
NM_001199159
|
PI16
|
peptidase inhibitor 16
|
|
chr17_+_18855477
|
0.059
|
NM_001042450
NM_152351
|
SLC5A10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10
|
|
chr15_-_40599872
|
0.058
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr8_-_145701139
|
0.057
|
|
FOXH1
|
forkhead box H1
|
|
chr14_-_94595925
|
0.057
|
NM_032036
|
IFI27L2
|
interferon, alpha-inducible protein 27-like 2
|
|
chr17_+_48133331
|
0.057
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr15_+_41523401
|
0.057
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr1_-_152009467
|
0.057
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr5_+_137673702
|
0.057
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr12_-_49351251
|
0.056
|
NM_001659
|
ARF3
|
ADP-ribosylation factor 3
|
|
chr12_-_42538432
|
0.056
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr11_+_9685650
|
0.056
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr12_+_90103470
|
0.055
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chrX_-_20284910
|
0.055
|
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr1_-_27930101
|
0.054
|
NM_001029882
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chr15_-_59041933
|
0.054
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr5_+_137673953
|
0.052
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr1_-_226926875
|
0.051
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr5_+_137673956
|
0.051
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr17_-_63052744
|
0.051
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr11_-_62474666
|
0.051
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr6_+_31590468
|
0.050
|
|
|
|
|
chr14_-_64009971
|
0.050
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr11_-_65487910
|
0.049
|
|
RNASEH2C
|
ribonuclease H2, subunit C
|