|
chr22_-_37640187
|
4.849
|
|
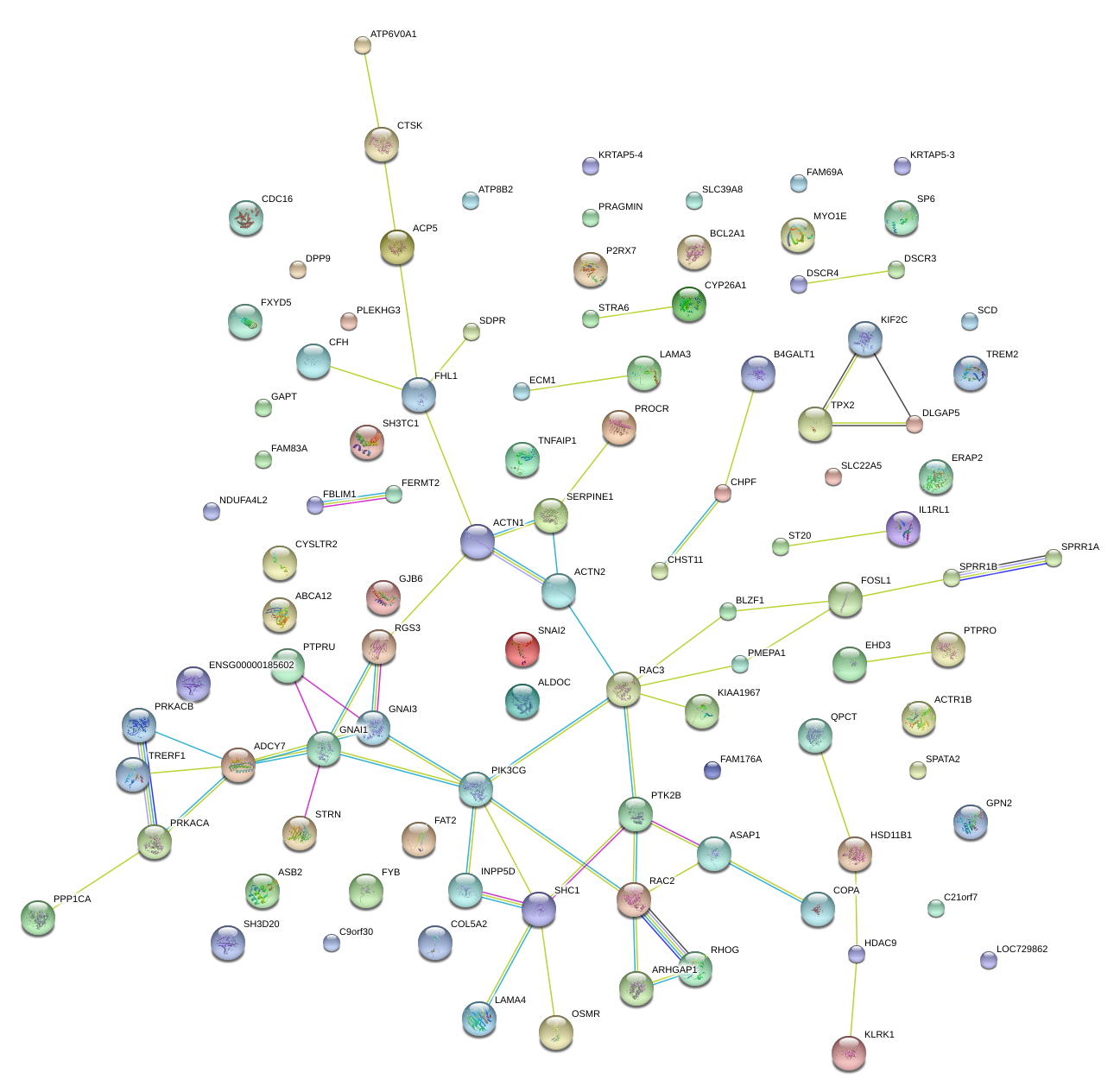
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr22_-_37640287
|
4.656
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr8_+_124194873
|
4.370
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr1_-_154946830
|
4.105
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr7_+_100770378
|
4.037
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr5_-_39219578
|
3.834
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr1_-_154946859
|
3.815
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr8_+_124194751
|
3.718
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr2_+_233925104
|
3.675
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr8_+_27182995
|
3.574
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr11_-_46722139
|
3.559
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr1_-_154946801
|
3.491
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr7_+_106505696
|
3.465
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr1_+_150480486
|
3.317
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr8_+_27183047
|
3.284
|
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr14_+_65171243
|
3.283
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr1_+_150480584
|
3.199
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr20_+_33759773
|
3.193
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr14_+_65171149
|
3.158
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr9_+_116356199
|
3.123
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr21_-_38639593
|
3.120
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr14_-_69446038
|
3.100
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr4_-_103266577
|
3.094
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr4_+_8200918
|
3.034
|
NM_018986
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1
|
|
chr2_+_233925035
|
2.986
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr5_-_39203061
|
2.886
|
|
FYB
|
FYN binding protein
|
|
chr14_-_55658205
|
2.868
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr11_-_46722160
|
2.856
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr2_+_31456879
|
2.854
|
NM_014600
|
EHD3
|
EH-domain containing 3
|
|
chr8_+_27183062
|
2.827
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr5_+_87564729
|
2.818
|
|
LOC100505894
|
uncharacterized LOC100505894
|
|
chr2_-_215896809
|
2.813
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr15_-_80263460
|
2.806
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr19_-_11689591
|
2.784
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr1_+_209859524
|
2.774
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr1_+_16083132
|
2.733
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr11_-_65667860
|
2.725
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr10_+_102107074
|
2.716
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr12_+_15699277
|
2.652
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr1_-_160313056
|
2.647
|
|
COPA
|
coatomer protein complex, subunit alpha
|
|
chr19_+_35645640
|
2.583
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr19_+_35645614
|
2.572
|
NM_014164
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr11_-_46722115
|
2.546
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr7_+_106505739
|
2.507
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chrX_+_135230736
|
2.499
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr17_+_26662547
|
2.447
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr10_+_102107256
|
2.411
|
|
|
|
|
chr16_+_50321822
|
2.398
|
NM_001114
|
ADCY7
|
adenylate cyclase 7
|
|
chr1_-_27216767
|
2.373
|
|
GPN2
|
GPN-loop GTPase 2
|
|
chr5_+_96212167
|
2.342
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr15_-_59665053
|
2.342
|
|
MYO1E
|
myosin IE
|
|
chr13_-_20805371
|
2.341
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr1_+_84609941
|
2.324
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr6_-_30652434
|
2.317
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr1_+_169337193
|
2.243
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr7_+_18548899
|
2.241
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr20_-_56286540
|
2.230
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr14_-_94442894
|
2.196
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr1_-_93342706
|
2.194
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr10_+_102106958
|
2.158
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr17_+_40610897
|
2.152
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr5_+_38845962
|
2.150
|
NM_003999
|
OSMR
|
oncostatin M receptor
|
|
chr1_-_27216585
|
2.137
|
|
GPN2
|
GPN-loop GTPase 2
|
|
chr1_-_160312959
|
2.129
|
|
|
|
|
chr1_+_110091185
|
2.124
|
NM_006496
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr14_-_94443065
|
2.123
|
NM_001202429
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr1_+_154300275
|
2.123
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr18_+_21452981
|
2.121
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr17_-_45933138
|
2.116
|
NM_199262
|
SP6
|
Sp6 transcription factor
|
|
chr17_+_40610940
|
2.107
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr2_+_102953716
|
2.079
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr8_-_49833823
|
2.076
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr7_+_106505876
|
2.072
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr13_+_49280619
|
2.070
|
NM_020377
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chr14_-_53417636
|
2.068
|
|
FERMT2
|
fermitin family member 2
|
|
chr1_-_150780705
|
2.066
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr8_-_131455833
|
2.051
|
NM_001247996
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr2_-_192711651
|
2.042
|
|
SDPR
|
serum deprivation response
|
|
chr11_-_1643324
|
2.005
|
NM_001012709
|
KRTAP5-4
|
keratin associated protein 5-4
|
|
chr2_-_190044330
|
2.003
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_-_39270708
|
2.003
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr2_-_75788027
|
1.993
|
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr2_+_37571752
|
1.987
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr15_-_74500394
|
1.980
|
NM_001199041
|
STRA6
|
stimulated by retinoic acid gene 6 homolog (mouse)
|
|
chr9_-_33167084
|
1.978
|
|
B4GALT1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1
|
|
chr2_-_75788079
|
1.975
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr12_-_123201319
|
1.970
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr21_+_30517897
|
1.967
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr10_+_94833646
|
1.945
|
NM_000783
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr8_-_8175860
|
1.945
|
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr9_+_103204187
|
1.912
|
NM_001198812
|
C9orf30-TMEFF1
C9orf30
|
C9orf30-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chr19_-_4723755
|
1.905
|
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr1_+_169337432
|
1.898
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr6_-_41130864
|
1.893
|
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr1_+_152956563
|
1.893
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr12_-_57630867
|
1.874
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr10_+_102107001
|
1.874
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr2_-_37193609
|
1.872
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr6_-_42419782
|
1.868
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr2_-_220407818
|
1.859
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr12_+_105153258
|
1.859
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr20_-_48532036
|
1.854
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr17_+_40610905
|
1.840
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr12_-_123187842
|
1.831
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr17_-_43510204
|
1.801
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr9_+_116356407
|
1.791
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr8_+_22462267
|
1.783
|
|
KIAA1967
|
KIAA1967
|
|
chr19_-_11688484
|
1.778
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr17_-_26903854
|
1.764
|
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr1_+_45205466
|
1.756
|
NM_006845
|
KIF2C
|
kinesin family member 2C
|
|
chr1_+_110091249
|
1.723
|
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr9_-_33167166
|
1.722
|
|
B4GALT1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1
|
|
chr20_+_30326903
|
1.714
|
NM_012112
|
TPX2
|
TPX2, microtubule-associated, homolog (Xenopus laevis)
|
|
chr5_+_57787260
|
1.703
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr19_-_4723791
|
1.699
|
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr11_+_844111
|
1.698
|
|
TSPAN4
|
tetraspanin 4
|
|
chr1_+_152881022
|
1.695
|
NM_005547
|
IVL
|
involucrin
|
|
chr14_-_100842637
|
1.692
|
|
WARS
|
tryptophanyl-tRNA synthetase
|
|
chr5_-_1634040
|
1.669
|
|
LOC728613
|
programmed cell death 6 pseudogene
|
|
chr4_-_10023094
|
1.668
|
NM_020041
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr2_-_136633934
|
1.662
|
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr7_+_143318541
|
1.662
|
NM_173678
|
LOC154761
FAM115C
|
uncharacterized LOC154761
family with sequence similarity 115, member C
|
|
chr19_-_4723814
|
1.657
|
NM_139159
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr5_-_171615095
|
1.656
|
|
STK10
|
serine/threonine kinase 10
|
|
chrX_-_153775774
|
1.650
|
NM_001042351
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr5_+_110074708
|
1.635
|
|
SLC25A46
|
solute carrier family 25, member 46
|
|
chr1_+_45205558
|
1.633
|
|
KIF2C
|
kinesin family member 2C
|
|
chr18_-_67872897
|
1.629
|
NM_173630
|
RTTN
|
rotatin
|
|
chr1_-_162381805
|
1.627
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr15_+_63340314
|
1.617
|
NM_001018008
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr10_+_114206988
|
1.612
|
|
VTI1A
|
vesicle transport through interaction with t-SNAREs homolog 1A (yeast)
|
|
chr20_-_23030141
|
1.607
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr8_-_42065193
|
1.605
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr16_+_77225094
|
1.604
|
|
MON1B
|
MON1 homolog B (yeast)
|
|
chr9_+_131185129
|
1.603
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr9_-_101017492
|
1.601
|
|
TBC1D2
|
TBC1 domain family, member 2
|
|
chr11_+_844062
|
1.599
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr16_+_1583657
|
1.597
|
|
TMEM204
|
transmembrane protein 204
|
|
chr6_-_41130921
|
1.596
|
NM_018965
|
TREM2
|
triggering receptor expressed on myeloid cells 2
|
|
chr2_-_216241321
|
1.595
|
|
FN1
|
fibronectin 1
|
|
chr19_+_41257086
|
1.593
|
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr17_+_9729380
|
1.588
|
NM_004246
|
GLP2R
|
glucagon-like peptide 2 receptor
|
|
chr3_-_50340979
|
1.588
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr1_+_45205533
|
1.587
|
|
KIF2C
|
kinesin family member 2C
|
|
chr7_-_100860991
|
1.576
|
NM_001084
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr12_+_48876285
|
1.576
|
NM_152319
|
C12orf54
|
chromosome 12 open reading frame 54
|
|
chr15_-_59665069
|
1.574
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr13_+_46039024
|
1.572
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr1_+_183114214
|
1.561
|
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr6_-_32160637
|
1.552
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr14_-_53417808
|
1.552
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr2_-_110371669
|
1.551
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr6_-_111888520
|
1.548
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_+_32688013
|
1.540
|
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I
|
|
chr1_+_192127591
|
1.539
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr2_-_161056573
|
1.534
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr1_-_27216784
|
1.534
|
NM_018066
|
GPN2
|
GPN-loop GTPase 2
|
|
chr6_-_131384321
|
1.533
|
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr1_-_109825683
|
1.531
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr2_+_239756672
|
1.526
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr1_+_32688010
|
1.524
|
|
EIF3I
|
eukaryotic translation initiation factor 3, subunit I
|
|
chr9_+_33750463
|
1.521
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr10_+_49892860
|
1.521
|
|
WDFY4
|
WDFY family member 4
|
|
chr7_+_86273867
|
1.519
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr5_+_68530713
|
1.515
|
|
CDK7
|
cyclin-dependent kinase 7
|
|
chr7_+_134464161
|
1.511
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr15_-_91565685
|
1.510
|
NM_018668
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast)
|
|
chr16_+_1583666
|
1.507
|
|
TMEM204
|
transmembrane protein 204
|
|
chr17_+_40610915
|
1.506
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr5_+_110074740
|
1.503
|
NM_138773
|
SLC25A46
|
solute carrier family 25, member 46
|
|
chr1_-_214724565
|
1.501
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr15_-_34610928
|
1.495
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr3_-_149095517
|
1.492
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr2_-_216259197
|
1.492
|
|
FN1
|
fibronectin 1
|
|
chr17_-_48262900
|
1.486
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr12_+_107712151
|
1.485
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr6_+_151561094
|
1.475
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr1_-_159923961
|
1.470
|
NM_001146172
NM_001146173
NM_033438
|
SLAMF9
|
SLAM family member 9
|
|
chr12_+_53442732
|
1.469
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr7_+_134464417
|
1.468
|
|
CALD1
|
caldesmon 1
|
|
chr15_-_41624760
|
1.458
|
NM_007280
|
OIP5
|
Opa interacting protein 5
|
|
chr20_+_44044706
|
1.455
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr7_+_143078608
|
1.454
|
|
ZYX
|
zyxin
|
|
chr7_+_129847699
|
1.452
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr10_-_71930185
|
1.450
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr17_-_39507055
|
1.450
|
NM_004138
|
KRT33A
|
keratin 33A
|
|
chr5_-_127873734
|
1.448
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr1_+_40506449
|
1.446
|
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|
|
chr12_-_47473424
|
1.444
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chr7_+_93551015
|
1.443
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr7_+_86274076
|
1.435
|
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr11_+_5617330
|
1.432
|
NM_058166
|
TRIM6
|
tripartite motif containing 6
|
|
chr5_+_68530621
|
1.432
|
NM_001799
|
CDK7
|
cyclin-dependent kinase 7
|
|
chr1_+_40506406
|
1.431
|
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast)
|
|
chr6_-_88411948
|
1.421
|
|
AKIRIN2
|
akirin 2
|
|
chr2_-_9563584
|
1.412
|
|
ITGB1BP1
|
integrin beta 1 binding protein 1
|
|
chr3_-_139258373
|
1.408
|
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr3_-_149095458
|
1.406
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chrX_-_153775427
|
1.404
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr15_+_69706619
|
1.400
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|