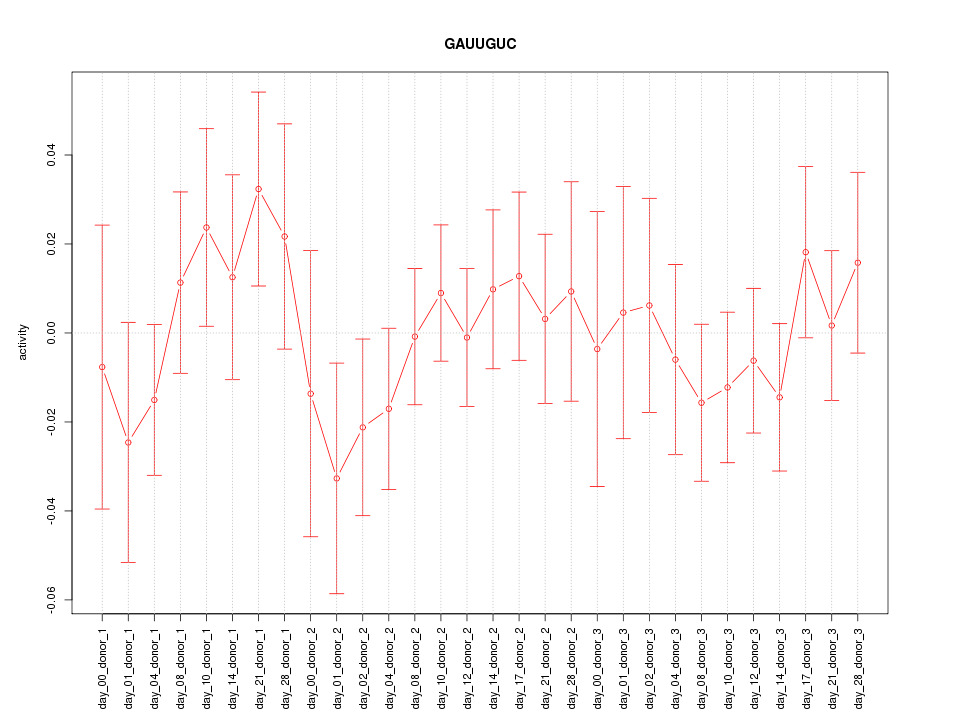
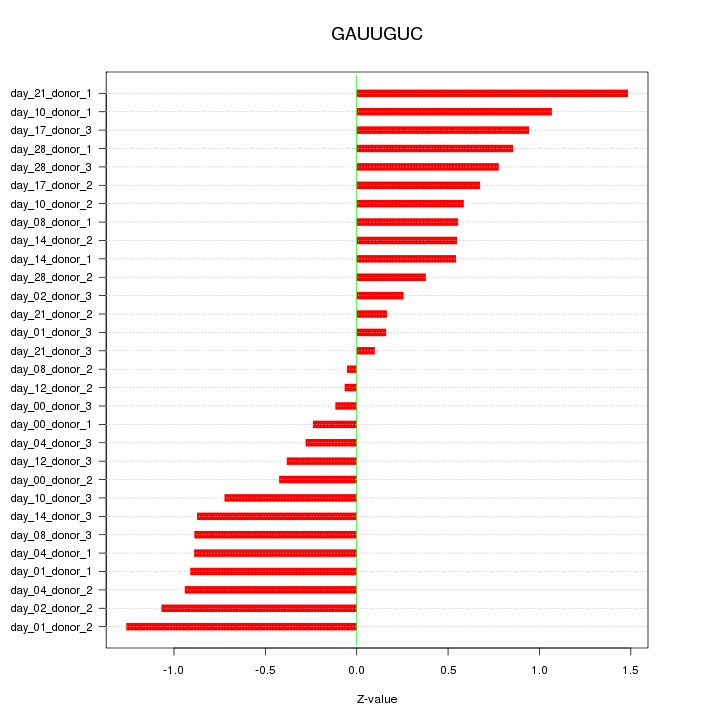
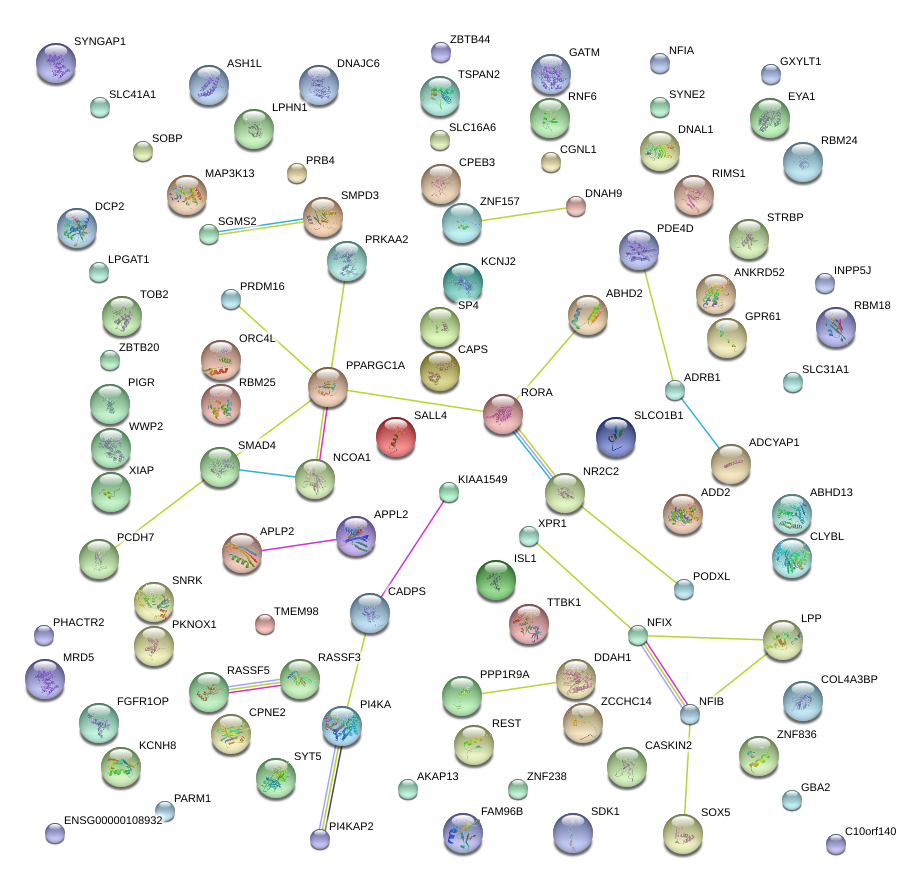
|
chr19_+_5914192
|
3.717
|
NM_004058
NM_080590
|
CAPS
|
calcyphosine
|
|
chr6_+_17281773
|
3.106
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282294
|
3.028
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282931
|
3.021
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr1_-_207119707
|
2.437
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr7_-_131241321
|
2.037
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr4_+_75858205
|
1.780
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr19_-_55691468
|
1.456
|
NM_003180
|
SYT5
|
synaptotagmin V
|
|
chr7_+_3340919
|
1.451
|
NM_152744
|
SDK1
|
sidekick cell adhesion molecule 1
|
|
chr1_+_244214552
|
1.372
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr5_+_50678957
|
1.321
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr14_+_74111577
|
1.272
|
NM_001201366
NM_031427
|
DNAL1
|
dynein, axonemal, light chain 1
|
|
chr1_+_244216506
|
1.203
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr15_-_60884615
|
0.974
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr7_+_21467572
|
0.960
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr19_-_14316980
|
0.955
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr1_-_86043932
|
0.896
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_85930733
|
0.886
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr15_-_61521478
|
0.876
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_-_115632047
|
0.841
|
NM_005725
|
TSPAN2
|
tetraspanin 2
|
|
chr1_+_180601121
|
0.833
|
NM_001135669
NM_004736
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr6_+_72926462
|
0.774
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr13_+_100258886
|
0.748
|
NM_206808
|
CLYBL
|
citrate lyase beta like
|
|
chr15_-_60919642
|
0.706
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_+_57668702
|
0.669
|
NM_001252335
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr9_-_35749209
|
0.665
|
NM_020944
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr1_+_61547625
|
0.651
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61542928
|
0.625
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_72268720
|
0.622
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr22_-_21088954
|
0.597
|
NM_002650
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr6_+_72922513
|
0.593
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr17_+_68165660
|
0.582
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr2_+_24807345
|
0.568
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr6_+_143999058
|
0.555
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_-_56652118
|
0.549
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr6_+_72926144
|
0.541
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr22_-_21213099
|
0.540
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr6_+_43211221
|
0.530
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr15_+_89631380
|
0.516
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr7_-_138665853
|
0.510
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr16_+_69958900
|
0.499
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr17_+_31254927
|
0.498
|
NM_001033504
NM_015544
|
TMEM98
|
transmembrane protein 98
|
|
chr22_+_31518923
|
0.493
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr13_+_108870713
|
0.487
|
NM_032859
|
ABHD13
|
abhydrolase domain containing 13
|
|
chr6_+_143929316
|
0.481
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr8_-_72274466
|
0.476
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr2_-_148779172
|
0.472
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr12_+_65004191
|
0.472
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr4_-_23891608
|
0.468
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr21_+_44394634
|
0.461
|
NM_004571
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr1_+_61547388
|
0.443
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr9_+_115983807
|
0.441
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr3_-_114343791
|
0.441
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr10_-_94050799
|
0.440
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr3_-_114343052
|
0.424
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr3_-_114478117
|
0.419
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_-_205782135
|
0.417
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr9_-_126030854
|
0.416
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr15_+_85923739
|
0.412
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr15_+_86220267
|
0.411
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr20_-_50418946
|
0.410
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr3_-_114102488
|
0.408
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_+_33387846
|
0.405
|
NM_006772
|
SYNGAP1
|
synaptic Ras GTPase activating protein 1
|
|
chr6_+_167412791
|
0.402
|
NM_007045
NM_194429
|
FGFR1OP
|
FGFR1 oncogene partner
|
|
chr14_+_64682671
|
0.393
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr9_-_126030792
|
0.389
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr3_+_185080835
|
0.388
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr6_+_73076431
|
0.382
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr2_-_148778262
|
0.379
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr14_+_73525187
|
0.379
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr3_+_185000714
|
0.378
|
NM_001242314
NM_001242317
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr15_-_45670955
|
0.365
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr1_+_110082493
|
0.362
|
NM_031936
|
GPR61
|
G protein-coupled receptor 61
|
|
chr3_+_43328001
|
0.361
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr3_-_114790221
|
0.358
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_105629790
|
0.357
|
NM_001251904
NM_018171
|
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2
|
|
chr12_-_42538432
|
0.355
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr22_-_41842853
|
0.354
|
NM_016272
|
TOB2
|
transducer of ERBB2, 2
|
|
chr1_-_155532322
|
0.353
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr1_+_57110745
|
0.352
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr12_-_24715349
|
0.350
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr3_+_14989076
|
0.349
|
NM_003298
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr3_+_187871473
|
0.340
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr4_+_57775064
|
0.339
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr13_-_26795927
|
0.334
|
NM_183043
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr14_+_64680858
|
0.334
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr3_+_187930720
|
0.334
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr4_+_108814419
|
0.329
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr13_-_26795821
|
0.326
|
NM_183044
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr9_-_125027080
|
0.322
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr4_+_108745718
|
0.320
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr18_+_48556483
|
0.318
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chr10_-_94003002
|
0.300
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chrX_+_122993536
|
0.296
|
NM_001204401
NM_001167
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr6_+_107811272
|
0.295
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr10_-_21814610
|
0.290
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr5_+_112312392
|
0.285
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr16_-_87525317
|
0.284
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr6_+_72596648
|
0.284
|
NM_014989
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_+_19189969
|
0.279
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr6_-_119031230
|
0.279
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr11_-_130184459
|
0.278
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr18_+_905296
|
0.270
|
NM_001117
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary)
|
|
chr1_+_2985730
|
0.269
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr17_-_66287256
|
0.262
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr16_-_68482373
|
0.257
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr1_+_65730423
|
0.255
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr10_+_115803805
|
0.255
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr14_+_64319607
|
0.254
|
NM_015180
NM_182914
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr4_+_30721865
|
0.250
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr3_-_114866090
|
0.249
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_74807448
|
0.243
|
NM_005713
NM_031361
|
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein
|
|
chr7_+_94536926
|
0.236
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr4_+_57774035
|
0.234
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|
|
chr1_-_212004113
|
0.228
|
NM_014873
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1
|
|
chr5_-_58334936
|
0.228
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_70994846
|
0.226
|
NM_001185054
|
ADD2
|
adducin 2 (beta)
|
|
chr5_-_74807805
|
0.222
|
NM_001130105
|
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein
|
|
chr16_+_57126439
|
0.219
|
NM_152727
|
CPNE2
|
copine II
|
|
chr12_-_12419702
|
0.216
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr12_-_23737507
|
0.214
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr15_+_52043738
|
0.209
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr7_+_94539209
|
0.203
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chrX_+_16964730
|
0.195
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr9_-_127905715
|
0.195
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr1_+_167189948
|
0.192
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_+_155089483
|
0.188
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr15_+_57210823
|
0.185
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr5_-_59783885
|
0.184
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_48343502
|
0.184
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr10_-_75173807
|
0.177
|
NM_001156
NM_004034
|
ANXA7
|
annexin A7
|
|
chr7_-_5463176
|
0.175
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr5_+_78531848
|
0.172
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr8_-_22550708
|
0.172
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr2_-_16847070
|
0.172
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr13_-_26796343
|
0.169
|
NM_005977
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr11_-_86666211
|
0.169
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr11_-_72852957
|
0.169
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr3_-_128206704
|
0.168
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr11_+_9595183
|
0.165
|
NM_003390
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr2_-_114514196
|
0.162
|
NM_025181
|
SLC35F5
|
solute carrier family 35, member F5
|
|
chr2_-_68547018
|
0.162
|
NM_001111101
NM_015463
|
CNRIP1
|
cannabinoid receptor interacting protein 1
|
|
chr1_+_167298280
|
0.159
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr5_+_142150291
|
0.155
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr14_-_93799371
|
0.153
|
NM_001002860
NM_018167
|
BTBD7
|
BTB (POZ) domain containing 7
|
|
chr8_+_17013799
|
0.152
|
NM_016353
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr12_+_72666462
|
0.146
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr6_-_38607923
|
0.145
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr12_-_49449106
|
0.142
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr6_-_86352635
|
0.139
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr19_+_1285766
|
0.139
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr1_-_31230673
|
0.137
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr15_+_64791618
|
0.136
|
NM_015042
|
ZNF609
|
zinc finger protein 609
|
|
chrX_-_54209639
|
0.135
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chrX_-_135338640
|
0.133
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr16_+_75032874
|
0.133
|
NM_032268
|
ZNRF1
|
zinc and ring finger 1
|
|
chr4_-_170924911
|
0.131
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr15_+_65822738
|
0.124
|
NM_016395
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1
|
|
chr8_+_132916339
|
0.124
|
NM_015137
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr6_-_109703703
|
0.123
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr15_-_23932410
|
0.120
|
NM_002487
|
NDN
|
necdin homolog (mouse)
|
|
chr6_-_107435635
|
0.120
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr11_-_6255891
|
0.119
|
NM_001098794
NM_032127
|
FAM160A2
|
family with sequence similarity 160, member A2
|
|
chr11_+_86511490
|
0.116
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr3_-_125094003
|
0.115
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr3_+_69812961
|
0.114
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr16_+_69796237
|
0.114
|
NM_007014
NM_199423
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr2_-_70995113
|
0.114
|
NM_001185055
NM_001617
NM_017482
NM_017488
|
ADD2
|
adducin 2 (beta)
|
|
chr19_-_47617008
|
0.112
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr12_+_72233486
|
0.112
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr8_+_61591239
|
0.110
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr6_-_38563763
|
0.109
|
NM_001172418
NM_152733
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr6_-_86352564
|
0.109
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr11_-_30038487
|
0.108
|
NM_002233
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4
|
|
chr3_+_30647901
|
0.107
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr2_-_60780404
|
0.106
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr5_-_56247915
|
0.106
|
NM_152622
|
MIER3
|
mesoderm induction early response 1, family member 3
|
|
chr6_-_118973019
|
0.106
|
NM_001042475
NM_206921
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr4_+_38665587
|
0.104
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr2_-_198175494
|
0.102
|
NM_001195144
NM_153697
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr5_+_65440662
|
0.102
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr11_-_124806302
|
0.100
|
NM_152722
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr3_+_29322802
|
0.100
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr3_+_69788562
|
0.099
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr12_+_94542239
|
0.098
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr9_-_88714486
|
0.097
|
NM_016548
|
GOLM1
|
golgi membrane protein 1
|
|
chr3_-_169381417
|
0.096
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr6_-_86351156
|
0.096
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_+_29562947
|
0.094
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr5_-_76934508
|
0.094
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr18_+_904943
|
0.093
|
NM_001099733
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary)
|
|
chr1_-_42800623
|
0.091
|
NM_001198851
|
FOXJ3
|
forkhead box J3
|
|
chr1_+_178995055
|
0.091
|
NM_014864
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr3_+_69915374
|
0.090
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_116164363
|
0.089
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr3_+_69812620
|
0.089
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_18466759
|
0.088
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr4_-_170947428
|
0.087
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr1_-_225840660
|
0.086
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr15_+_57511616
|
0.085
|
NM_207040
|
TCF12
|
transcription factor 12
|