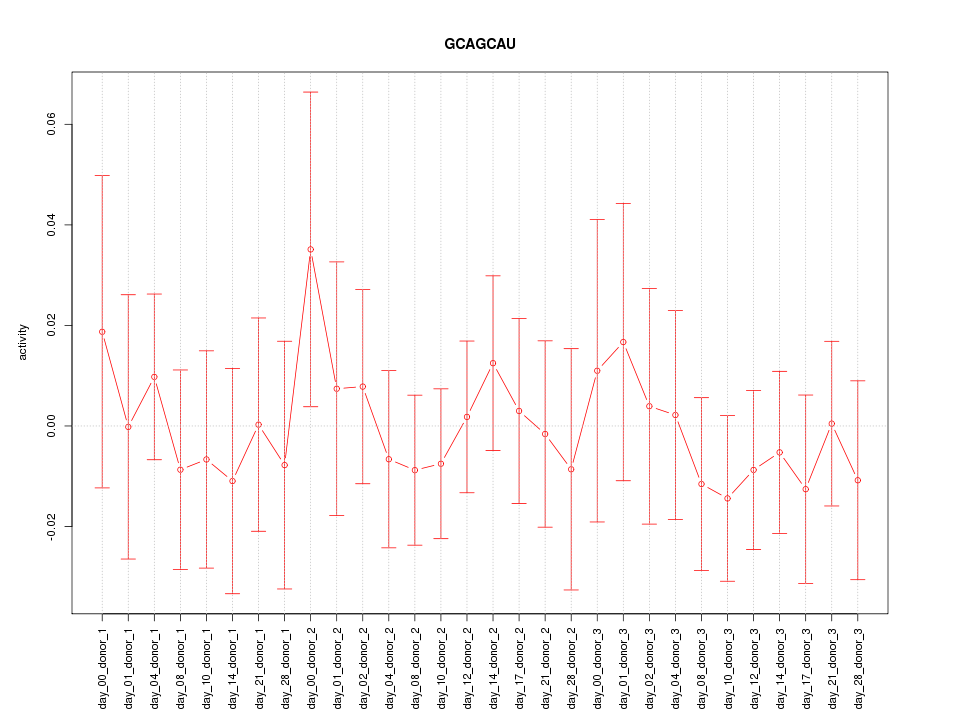
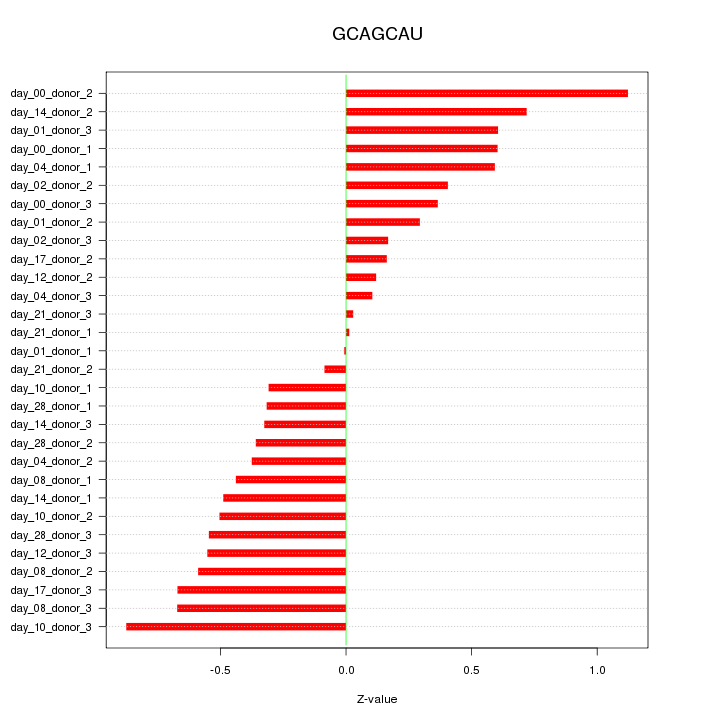
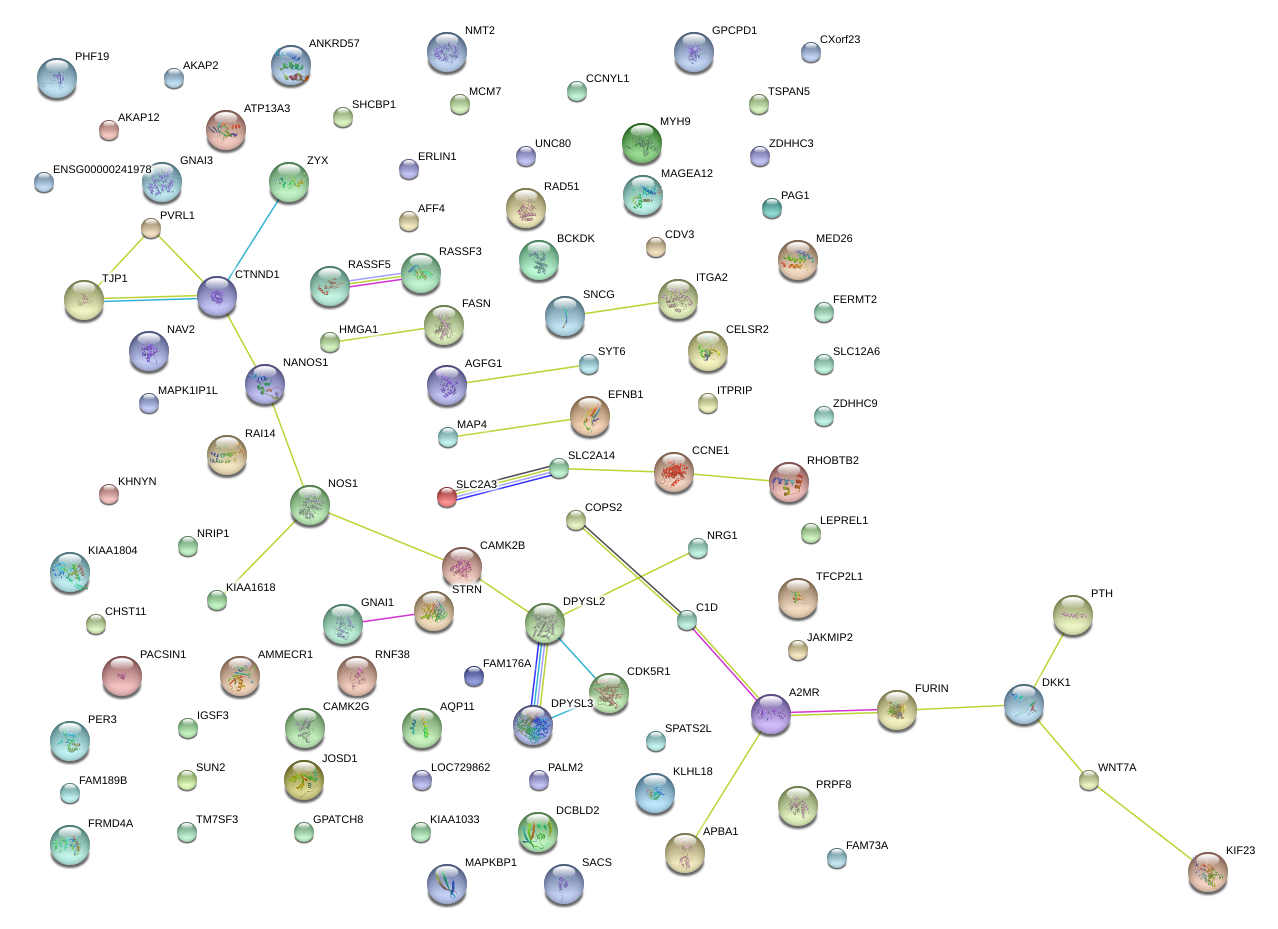
|
chr6_+_151646634
|
1.293
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_+_151561094
|
1.219
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr15_+_69706619
|
1.002
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chr10_+_88718287
|
0.974
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr3_-_98620452
|
0.890
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr6_+_34204576
|
0.826
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_+_34204647
|
0.815
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_+_34206567
|
0.810
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr4_-_99579540
|
0.797
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr9_-_72287097
|
0.794
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr12_-_117747214
|
0.745
|
NM_001204214
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr2_+_208576239
|
0.728
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr16_-_46655303
|
0.689
|
NM_024745
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr15_-_34629941
|
0.645
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr12_-_117742856
|
0.638
|
NM_001204213
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr15_-_34630203
|
0.636
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr10_-_15210610
|
0.615
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr1_+_109792640
|
0.594
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr21_-_16437125
|
0.592
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr15_-_34629044
|
0.571
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr15_-_34610928
|
0.551
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr9_+_112887780
|
0.549
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr10_-_75634219
|
0.542
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr22_-_36784055
|
0.517
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr12_-_117799410
|
0.506
|
NM_000620
NM_001204218
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr8_-_82024277
|
0.499
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr20_-_5591487
|
0.495
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr12_-_8088629
|
0.487
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr3_-_189840225
|
0.479
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr9_+_112810877
|
0.478
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chrX_-_109683457
|
0.462
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr10_-_106093662
|
0.450
|
NM_033397
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein
|
|
chr11_+_77300679
|
0.447
|
NM_173039
|
AQP11
|
aquaporin 11
|
|
chr8_+_26371461
|
0.444
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_48130633
|
0.435
|
NM_001134364
NM_002375
NM_030885
|
MAP4
|
microtubule-associated protein 4
|
|
chr22_-_39095991
|
0.433
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr12_+_57522247
|
0.432
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chrX_-_109561314
|
0.424
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr17_-_80056104
|
0.423
|
NM_004104
|
FASN
|
fatty acid synthase
|
|
chr5_+_34684611
|
0.415
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr1_+_110091185
|
0.412
|
NM_006496
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3
|
|
chr10_+_54074037
|
0.408
|
NM_012242
|
DKK1
|
dickkopf 1 homolog (Xenopus laevis)
|
|
chr3_-_45017600
|
0.398
|
NM_001135179
NM_016598
|
ZDHHC3
|
zinc finger, DHHC-type containing 3
|
|
chr12_+_105501448
|
0.390
|
NM_015275
|
KIAA1033
|
KIAA1033
|
|
chr17_+_30813928
|
0.384
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr8_+_26435339
|
0.380
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_189838907
|
0.380
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr15_+_42066631
|
0.379
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr10_-_14372807
|
0.378
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr2_+_228336877
|
0.365
|
NM_001135187
NM_001135188
NM_001135189
NM_004504
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr1_+_206680863
|
0.364
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr22_-_39151457
|
0.358
|
NM_001199580
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr7_+_143078358
|
0.357
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr3_-_13921617
|
0.357
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chrX_+_68048792
|
0.355
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr11_+_19372270
|
0.350
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr1_-_117210301
|
0.347
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr5_-_147162251
|
0.336
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr2_+_201170603
|
0.335
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr2_-_37193609
|
0.334
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr15_+_91411884
|
0.332
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr8_+_31497267
|
0.331
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr1_+_78245306
|
0.321
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr5_+_34687663
|
0.315
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr12_-_27167332
|
0.314
|
NM_016551
|
TM7SF3
|
transmembrane 7 superfamily member 3
|
|
chr22_-_39152008
|
0.312
|
NM_001199579
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr5_-_146889618
|
0.311
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr5_+_34656595
|
0.308
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr8_+_22853360
|
0.307
|
NM_001160037
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr5_-_132299263
|
0.306
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr19_+_30302861
|
0.302
|
NM_001238
|
CCNE1
|
cyclin E1
|
|
chr5_+_34656367
|
0.300
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr11_-_119599253
|
0.295
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr6_+_34482646
|
0.292
|
NM_001199583
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr3_-_194188967
|
0.289
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr10_-_101945686
|
0.288
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr3_+_47324329
|
0.288
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr15_-_30114536
|
0.287
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr14_+_24899139
|
0.286
|
NM_015299
|
KHNYN
|
KH and NYN domain containing
|
|
chr2_-_75796847
|
0.284
|
NM_032181
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr11_+_19734821
|
0.283
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr1_-_155225137
|
0.281
|
NM_006589
NM_198264
|
FAM189B
|
family with sequence similarity 189, member B
|
|
chr8_+_32504250
|
0.279
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr11_-_13517525
|
0.278
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr1_-_114695061
|
0.277
|
NM_205848
|
SYT6
|
synaptotagmin VI
|
|
chr7_-_99699426
|
0.277
|
NM_005916
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr14_-_53417808
|
0.277
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr13_-_24007822
|
0.276
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr5_-_146833150
|
0.276
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr10_-_101945787
|
0.274
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr1_+_206730456
|
0.273
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr3_+_133293256
|
0.271
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr2_+_210636700
|
0.270
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chr9_-_36400212
|
0.269
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chr17_-_42580927
|
0.267
|
NM_001002909
|
GPATCH8
|
G patch domain containing 8
|
|
chr2_-_122042767
|
0.266
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr9_-_123639566
|
0.266
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr8_+_22857063
|
0.265
|
NM_015178
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr11_+_57529233
|
0.264
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr2_+_201170795
|
0.261
|
NM_001100422
NM_001100424
NM_001100423
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr1_+_233463513
|
0.258
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chrX_-_128977291
|
0.257
|
NM_001008222
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr19_-_16738986
|
0.256
|
NM_004831
|
MED26
|
mediator complex subunit 26
|
|
chr1_+_7844713
|
0.251
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr9_+_112542496
|
0.251
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr16_+_31119661
|
0.249
|
NM_001122957
NM_005881
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|
|
chr12_+_104850641
|
0.248
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr16_-_88851371
|
0.246
|
NM_001142864
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1
|
|
chr7_-_99698278
|
0.246
|
NM_182776
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr14_+_55518313
|
0.246
|
NM_144578
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like
|
|
chr15_-_49447765
|
0.242
|
NM_001143887
NM_004236
|
COPS2
|
COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis)
|
|
chr15_+_40987326
|
0.242
|
NM_001164270
NM_002875
NM_133487
NM_001164269
|
RAD51
|
RAD51 homolog (S. cerevisiae)
|
|
chrX_-_19988381
|
0.241
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr5_+_52284903
|
0.241
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr2_+_110371887
|
0.237
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr17_+_8339135
|
0.236
|
NM_001025579
NM_030808
|
NDEL1
|
nudE nuclear distribution gene E homolog (A. nidulans)-like 1
|
|
chrX_-_106959597
|
0.235
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr15_-_63674074
|
0.235
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr4_+_4387982
|
0.232
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr1_-_29450406
|
0.232
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr6_-_11382501
|
0.229
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_99873129
|
0.229
|
NM_015491
NM_032870
|
PNISR
|
PNN-interacting serine/arginine-rich protein
|
|
chr4_-_153303663
|
0.228
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr9_-_36400817
|
0.227
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chr2_-_165697927
|
0.224
|
NM_014900
|
COBLL1
|
COBL-like 1
|
|
chrX_-_41782230
|
0.223
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr1_-_32169619
|
0.221
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chrX_-_128977719
|
0.216
|
NM_016032
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr2_+_113875469
|
0.215
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr13_-_21635703
|
0.214
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr2_-_71062904
|
0.213
|
NM_015717
|
CD207
|
CD207 molecule, langerin
|
|
chrX_-_48815641
|
0.212
|
NM_001136159
|
OTUD5
|
OTU domain containing 5
|
|
chr2_-_46384
|
0.210
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr2_-_69870976
|
0.207
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr1_-_29449012
|
0.207
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr1_-_41131323
|
0.205
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr10_+_75757847
|
0.204
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr15_+_93443418
|
0.204
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr12_-_53074172
|
0.204
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr1_-_179198814
|
0.202
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr11_-_27681195
|
0.200
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr10_+_101419169
|
0.200
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr1_-_240775448
|
0.196
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr2_-_75788079
|
0.196
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr7_-_121944485
|
0.195
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr2_-_206950895
|
0.193
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr7_-_92463211
|
0.193
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr9_+_33817168
|
0.192
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr1_-_171711093
|
0.191
|
NM_001185127
NM_003762
|
VAMP4
|
vesicle-associated membrane protein 4
|
|
chr11_-_16497917
|
0.191
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr2_-_171923482
|
0.189
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr8_-_80993009
|
0.189
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chrX_-_131351917
|
0.189
|
NM_021183
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr8_-_81083660
|
0.187
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr6_+_34433849
|
0.186
|
NM_020804
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr8_+_59465727
|
0.185
|
NM_001007067
NM_001007068
NM_001007069
NM_001007070
NM_005625
|
SDCBP
|
syndecan binding protein (syntenin)
|
|
chr2_+_113885137
|
0.185
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr10_-_64028457
|
0.184
|
NM_145307
|
RTKN2
|
rhotekin 2
|
|
chr16_-_48644087
|
0.184
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr17_-_45266664
|
0.184
|
NM_001114091
NM_001256
|
CDC27
|
cell division cycle 27 homolog (S. cerevisiae)
|
|
chr5_-_149682447
|
0.184
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr12_+_49760679
|
0.184
|
NM_023071
|
SPATS2
|
spermatogenesis associated, serine-rich 2
|
|
chr14_+_33408458
|
0.183
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr22_-_28197469
|
0.183
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr12_+_26205490
|
0.181
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr10_+_52750904
|
0.181
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr10_+_103113807
|
0.177
|
NM_003939
NM_033637
|
BTRC
|
beta-transducin repeat containing
|
|
chr1_+_36273786
|
0.174
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr10_+_95653668
|
0.170
|
NM_001134658
NM_153226
|
SLC35G1
|
solute carrier family 35, member G1
|
|
chr17_+_7554239
|
0.169
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr1_-_53793743
|
0.169
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr22_+_32340435
|
0.167
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr3_+_151985828
|
0.167
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr12_+_57943843
|
0.166
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr11_+_111473099
|
0.165
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chrX_-_106960268
|
0.165
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chrX_+_134478671
|
0.164
|
NM_152695
|
ZNF449
|
zinc finger protein 449
|
|
chr7_+_28338939
|
0.162
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_-_93075190
|
0.162
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_150849211
|
0.162
|
NM_001197325
NM_001668
NM_178427
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator
|
|
chr1_+_201708940
|
0.162
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr2_-_131850988
|
0.161
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr20_+_24449834
|
0.161
|
NM_024893
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr11_+_76571902
|
0.160
|
NM_018367
|
ACER3
|
alkaline ceramidase 3
|
|
chr19_-_44285407
|
0.160
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_-_214724974
|
0.158
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr2_-_29297126
|
0.157
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr7_+_128379345
|
0.157
|
NM_001130674
NM_001199671
NM_001199672
NM_001199673
NM_001199674
NM_001219
|
CALU
|
calumenin
|
|
chr16_+_18995255
|
0.157
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chrX_+_30671456
|
0.156
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr1_-_67520028
|
0.155
|
NM_015139
|
SLC35D1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1
|
|
chr7_-_82073020
|
0.153
|
NM_000722
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chrX_-_107019001
|
0.153
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr22_-_42017032
|
0.152
|
NM_015704
|
PPPDE2
|
PPPDE peptidase domain containing 2
|
|
chr4_+_124317884
|
0.152
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_-_42801529
|
0.152
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chr6_+_47666288
|
0.151
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr19_-_17559375
|
0.150
|
NM_001190844
|
TMEM221
|
transmembrane protein 221
|
|
chr8_-_125740650
|
0.150
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr3_-_122233781
|
0.149
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|