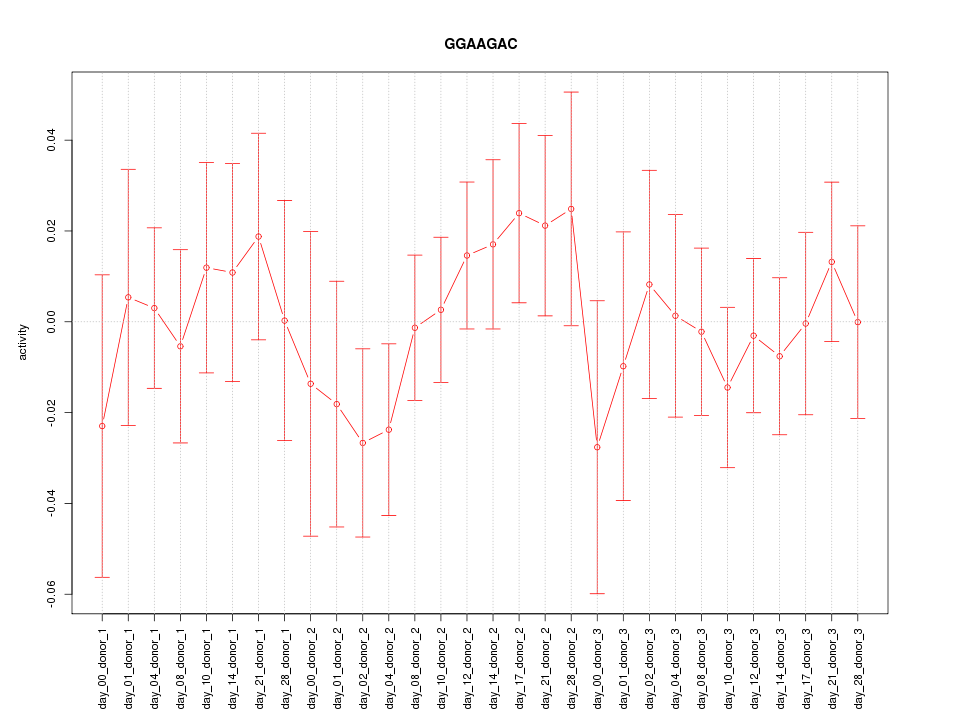
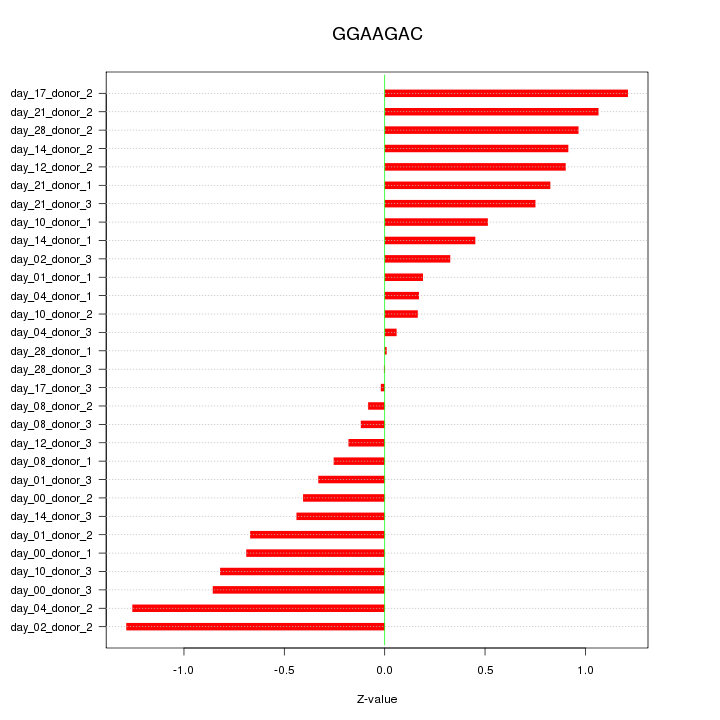
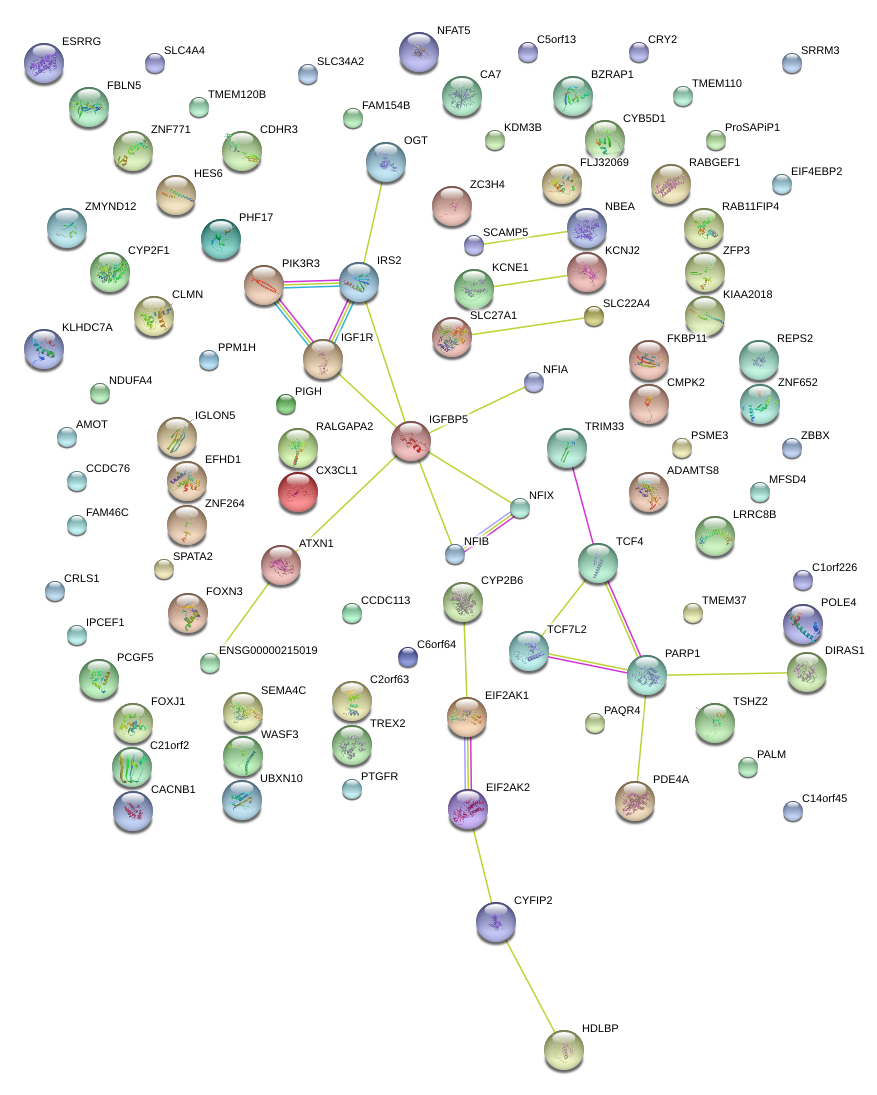
|
chr4_+_72052354
|
1.365
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_-_217262946
|
1.119
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_217311095
|
1.068
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr14_-_90085325
|
1.057
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr1_-_216896640
|
1.040
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr17_-_56406120
|
1.027
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr14_-_89883306
|
0.997
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr1_+_61547625
|
0.976
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_61542928
|
0.971
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr17_+_68165660
|
0.951
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr1_+_162351519
|
0.930
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr19_+_10543110
|
0.837
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr13_+_115079943
|
0.817
|
NM_001164144
NM_001164145
NM_032436
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr1_+_61547388
|
0.802
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr4_+_72204769
|
0.735
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr19_+_41620336
|
0.725
|
NM_000774
|
CYP2F1
|
cytochrome P450, family 2, subfamily F, polypeptide 1
|
|
chr1_+_118148506
|
0.700
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr19_+_10563581
|
0.700
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr1_-_46598250
|
0.696
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr19_+_10531128
|
0.688
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_+_10541334
|
0.638
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr11_+_45868956
|
0.612
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr20_+_5986560
|
0.612
|
NM_019095
|
CRLS1
|
cardiolipin synthase 1
|
|
chr2_+_120189342
|
0.589
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr2_+_75185765
|
0.581
|
NM_019896
|
POLE4
|
polymerase (DNA-directed), epsilon 4 (p12 subunit)
|
|
chr11_+_45868668
|
0.578
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr5_-_111093251
|
0.575
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr20_-_20693122
|
0.571
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr5_-_111093030
|
0.571
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_47439326
|
0.557
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr5_-_111092918
|
0.550
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_111093792
|
0.548
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_75831194
|
0.540
|
NM_001110199
|
SRRM3
|
serine/arginine repetitive matrix 3
|
|
chr20_+_5987897
|
0.525
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr1_+_89990396
|
0.522
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr2_-_55459406
|
0.522
|
NM_152385
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chr17_-_74137370
|
0.508
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr17_+_29718641
|
0.502
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr4_+_129730772
|
0.496
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr2_-_55459698
|
0.495
|
NM_001135598
|
C2orf63
|
chromosome 2 open reading frame 63
|
|
chr1_-_42921923
|
0.494
|
NM_001146192
NM_032257
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr5_+_137688270
|
0.489
|
NM_016604
|
KDM3B
|
lysine (K)-specific demethylase 3B
|
|
chr14_-_68066919
|
0.482
|
NM_004569
|
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H
|
|
chr21_-_35831856
|
0.479
|
NM_001127668
NM_001127669
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chrX_-_112066353
|
0.478
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr21_-_35883570
|
0.474
|
NM_000219
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr1_+_100598705
|
0.473
|
NM_019083
|
CCDC76
|
coiled-coil domain containing 76
|
|
chr13_-_110438896
|
0.463
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr3_-_52931595
|
0.459
|
NM_001198974
NM_198563
|
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough
transmembrane protein 110
|
|
chr1_+_78956727
|
0.456
|
NM_000959
NM_001039585
|
PTGFR
|
prostaglandin F receptor (FP)
|
|
chr17_+_40985416
|
0.454
|
NM_005789
NM_176863
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr20_-_48530275
|
0.454
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr21_-_35828038
|
0.449
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr5_-_111312522
|
0.442
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr19_+_51815095
|
0.438
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr13_+_27131799
|
0.406
|
NM_006646
|
WASF3
|
WAS protein family, member 3
|
|
chr18_-_53255734
|
0.402
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chrX_+_16964730
|
0.391
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr1_+_90024271
|
0.388
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr14_-_92414004
|
0.387
|
NM_006329
|
FBLN5
|
fibulin 5
|
|
chrX_+_70752911
|
0.386
|
NM_181672
NM_181673
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr3_-_167098070
|
0.376
|
NM_001199202
NM_001199201
NM_024687
|
ZBBX
|
zinc finger, B-box domain containing
|
|
chr12_-_63328663
|
0.368
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr15_+_75287829
|
0.367
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr2_-_217560247
|
0.359
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr10_+_72163860
|
0.357
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr7_+_66093854
|
0.356
|
NM_001167961
NM_153033
|
KCTD7
|
potassium channel tetramerisation domain containing 7
|
|
chr1_-_115053758
|
0.355
|
NM_015906
NM_033020
|
TRIM33
|
tripartite motif containing 33
|
|
chr16_+_58283819
|
0.355
|
NM_001142302
NM_014157
|
CCDC113
|
coiled-coil domain containing 113
|
|
chr14_-_95786095
|
0.354
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr2_-_242212244
|
0.345
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr19_-_47617008
|
0.345
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr6_-_16761600
|
0.338
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr7_+_105603656
|
0.335
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr12_+_122150638
|
0.335
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr15_+_99192696
|
0.330
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr16_+_66878281
|
0.328
|
NM_005182
|
CA7
|
carbonic anhydrase VII
|
|
chr5_+_131630065
|
0.319
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr3_-_113415313
|
0.315
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr5_+_156693051
|
0.315
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr20_+_51588945
|
0.310
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chrX_-_112084006
|
0.308
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr1_-_226595796
|
0.302
|
NM_001618
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr6_-_154677868
|
0.302
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr19_+_708766
|
0.302
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr5_+_156696351
|
0.296
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr15_+_82555101
|
0.296
|
NM_001008226
|
FAM154B
|
family with sequence similarity 154, member B
|
|
chr1_+_205538111
|
0.295
|
NM_181644
|
MFSD4
|
major facilitator superfamily domain containing 4
|
|
chr2_-_239148626
|
0.295
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr19_+_41497182
|
0.292
|
NM_000767
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr7_-_6098813
|
0.291
|
NM_001134335
NM_014413
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1
|
|
chr2_+_233498206
|
0.291
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chr7_-_10979752
|
0.287
|
NM_002489
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
|
|
chr20_+_51801821
|
0.287
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr19_-_2721337
|
0.285
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr4_+_25658085
|
0.283
|
NM_001177999
|
SLC34A2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr6_-_154651214
|
0.281
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr16_+_57406398
|
0.277
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr13_+_36050885
|
0.277
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr20_-_3149070
|
0.276
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr1_+_20512577
|
0.273
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr16_+_3019341
|
0.269
|
NM_152341
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr6_-_39082750
|
0.269
|
NM_018322
|
C6orf64
|
chromosome 6 open reading frame 64
|
|
chr10_+_92980322
|
0.269
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr2_-_7005764
|
0.265
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr1_+_18807423
|
0.261
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr21_-_45759268
|
0.259
|
NM_004928
|
C21orf2
|
chromosome 21 open reading frame 2
|
|
chr16_+_69599868
|
0.257
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr14_+_74486049
|
0.257
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chr19_+_57702833
|
0.255
|
NM_003417
|
ZNF264
|
zinc finger protein 264
|
|
chr2_-_97535669
|
0.253
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr17_+_4981721
|
0.253
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr17_+_7761063
|
0.251
|
NM_144607
|
CYB5D1
|
cytochrome b5 domain containing 1
|
|
chr16_+_19467771
|
0.251
|
NM_024780
|
TMC5
|
transmembrane channel-like 5
|
|
chr22_-_39548448
|
0.250
|
NM_175709
|
CBX7
|
chromobox homolog 7
|
|
chr17_+_5390212
|
0.250
|
NM_024039
|
MIS12
|
MIS12, MIND kinetochore complex component, homolog (S. pombe)
|
|
chr12_+_133757994
|
0.250
|
NM_001165883
NM_001165884
NM_001165885
NM_001165886
NM_001165887
NM_003415
NM_152943
NM_001165881
NM_001165882
|
ZNF268
|
zinc finger protein 268
|
|
chr9_-_99329092
|
0.249
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr16_+_66878840
|
0.249
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chrX_+_150565655
|
0.248
|
NM_001017980
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae)
|
|
chr7_+_2557496
|
0.242
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr8_+_17354562
|
0.242
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr14_+_58765209
|
0.241
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr1_+_212738675
|
0.241
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr16_-_18937722
|
0.240
|
NM_015092
|
SMG1
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr21_-_43735705
|
0.240
|
NM_003226
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr18_+_9137560
|
0.239
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr1_+_162467594
|
0.238
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr9_-_4300028
|
0.237
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr13_+_35516390
|
0.236
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr4_+_25657434
|
0.236
|
NM_001177998
NM_006424
|
SLC34A2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr15_+_57210823
|
0.234
|
NM_003205
NM_207036
NM_207037
NM_207038
|
TCF12
|
transcription factor 12
|
|
chr10_+_96305521
|
0.233
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr17_-_9929567
|
0.231
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr1_+_212781969
|
0.230
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr10_-_94333847
|
0.229
|
NM_004969
|
IDE
|
insulin-degrading enzyme
|
|
chr7_+_2552162
|
0.227
|
NM_001166355
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr9_-_4152182
|
0.226
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr12_+_65218351
|
0.226
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr8_+_104152876
|
0.225
|
NM_001024372
NM_024812
|
BAALC
|
brain and acute leukemia, cytoplasmic
|
|
chr16_-_12010518
|
0.225
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr6_+_84743250
|
0.223
|
NM_138409
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr5_+_149109814
|
0.220
|
NM_001172698
NM_133263
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr7_-_112430408
|
0.217
|
NM_022484
|
TMEM168
|
transmembrane protein 168
|
|
chr7_+_43966007
|
0.217
|
NM_015983
|
UBE2D4
|
ubiquitin-conjugating enzyme E2D 4 (putative)
|
|
chr15_-_40398286
|
0.214
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr3_-_88108047
|
0.214
|
NM_001008390
NM_003663
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr5_+_149151502
|
0.212
|
NM_001172699
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_+_162466963
|
0.212
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr15_-_40398573
|
0.211
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr8_+_75896707
|
0.211
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr17_-_9940004
|
0.207
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr13_-_20110884
|
0.206
|
NM_001141968
NM_130785
NM_199254
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2
|
|
chr5_-_111091947
|
0.206
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_10101854
|
0.205
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr15_+_43809805
|
0.204
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr17_+_42977141
|
0.203
|
NM_213607
|
CCDC103
|
coiled-coil domain containing 103
|
|
chr9_-_99382073
|
0.202
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr1_+_45965855
|
0.200
|
NM_015506
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria
|
|
chr20_+_30945988
|
0.200
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr8_+_17396285
|
0.199
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr11_-_119234859
|
0.197
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr16_+_19422056
|
0.196
|
NM_001105248
NM_001105249
|
TMC5
|
transmembrane channel-like 5
|
|
chr1_+_183605207
|
0.195
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr16_-_4166185
|
0.194
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr15_+_44084039
|
0.194
|
NM_001018108
NM_001199875
NM_001199876
|
SERF2
|
small EDRK-rich factor 2
|
|
chr10_-_17244069
|
0.194
|
NM_004412
|
TRDMT1
|
tRNA aspartic acid methyltransferase 1
|
|
chr6_+_90272036
|
0.192
|
NM_001242811
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr1_+_36348795
|
0.190
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr20_-_30310597
|
0.186
|
NM_001191
NM_138578
|
BCL2L1
|
BCL2-like 1
|
|
chr2_+_97426571
|
0.186
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chrX_-_53449610
|
0.185
|
NM_006306
|
SMC1A
|
structural maintenance of chromosomes 1A
|
|
chr19_+_11925077
|
0.184
|
NM_152357
|
ZNF440
|
zinc finger protein 440
|
|
chr6_-_33285718
|
0.183
|
NM_001145338
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr12_-_50297575
|
0.180
|
NM_012306
|
FAIM2
|
Fas apoptotic inhibitory molecule 2
|
|
chr17_-_4167028
|
0.180
|
NM_016376
NM_020740
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr18_+_44526786
|
0.179
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr15_-_40401041
|
0.179
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr4_-_23891608
|
0.177
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr6_-_33285493
|
0.177
|
NM_005453
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr7_+_138818489
|
0.177
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr15_+_44069272
|
0.173
|
NM_001199877
|
SERF2
|
small EDRK-rich factor 2
|
|
chr10_+_81838417
|
0.172
|
NM_025125
|
FAM213A
|
family with sequence similarity 213, member A
|
|
chr5_-_58334936
|
0.171
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr14_-_94856926
|
0.171
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr11_-_70507911
|
0.170
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr15_+_78556901
|
0.170
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_+_78558522
|
0.169
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr16_+_14013989
|
0.168
|
NM_005236
|
ERCC4
|
excision repair cross-complementing rodent repair deficiency, complementation group 4
|
|
chr14_-_94855120
|
0.167
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr15_-_54051837
|
0.165
|
NM_182758
|
WDR72
|
WD repeat domain 72
|
|
chr6_-_80247103
|
0.165
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr5_+_127419407
|
0.164
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr11_+_101785720
|
0.163
|
NM_020802
|
KIAA1377
|
KIAA1377
|
|
chr1_-_207095198
|
0.163
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr17_-_33700619
|
0.163
|
NM_001104587
NM_001104588
NM_001104589
NM_001104590
NM_152270
|
SLFN11
|
schlafen family member 11
|
|
chr15_-_68498373
|
0.162
|
NM_001031733
NM_033429
|
CALML4
|
calmodulin-like 4
|
|
chr6_+_90142802
|
0.161
|
NM_001242809
NM_001242813
NM_014942
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr6_+_138483024
|
0.161
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr20_-_33460558
|
0.160
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|