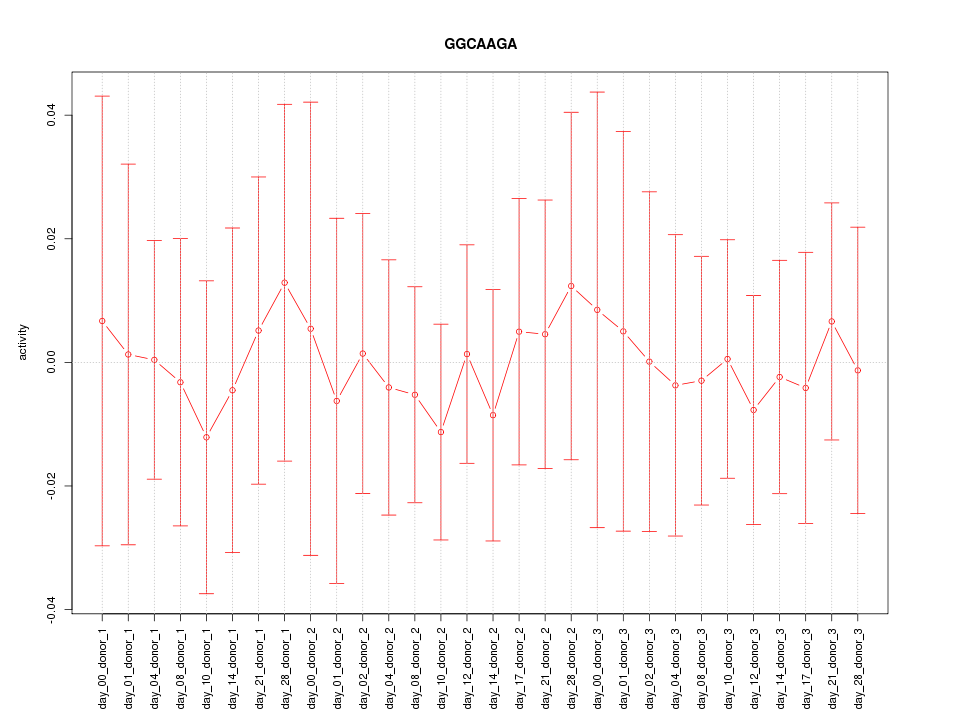
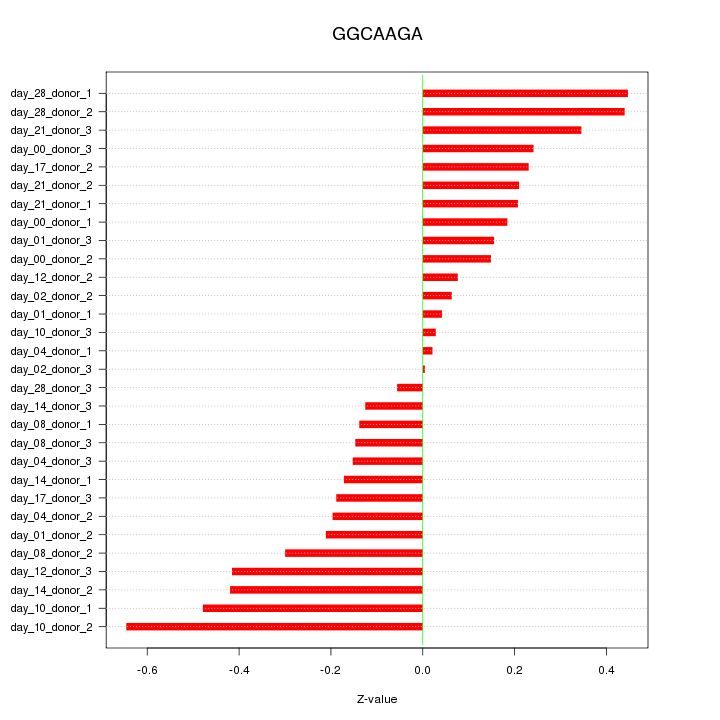
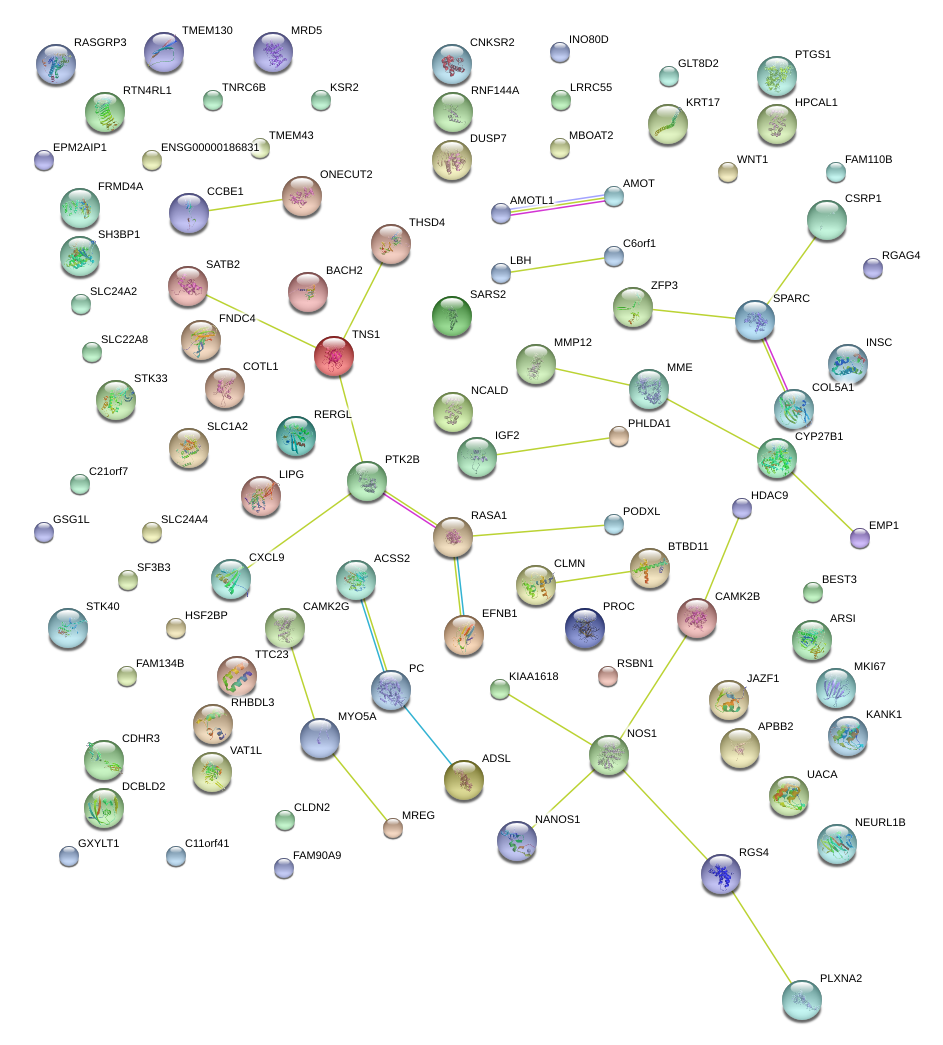
|
chr11_-_66725790
|
0.183
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr11_-_66675334
|
0.171
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr4_-_41216455
|
0.128
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr3_-_52090090
|
0.126
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr4_-_40859081
|
0.110
|
NM_001166051
NM_001166052
NM_001166053
NM_001166054
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr16_+_70557688
|
0.108
|
NM_012426
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr15_-_52821004
|
0.104
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr5_+_60933635
|
0.099
|
NM_173667
|
C5orf64
|
chromosome 5 open reading frame 64
|
|
chr6_-_91006460
|
0.093
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_-_91006580
|
0.092
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr11_+_56949220
|
0.092
|
NM_001005210
|
LRRC55
|
leucine rich repeat containing 55
|
|
chr5_+_86564727
|
0.091
|
NM_022650
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr9_+_706744
|
0.091
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr15_-_71055745
|
0.090
|
NM_018003
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr2_-_9143742
|
0.086
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr12_-_117747214
|
0.086
|
NM_001204214
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr12_-_76425375
|
0.083
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr5_+_86564044
|
0.082
|
NM_002890
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1
|
|
chr2_-_200335988
|
0.081
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr9_+_504661
|
0.081
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr9_+_137533643
|
0.079
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr11_-_35441099
|
0.076
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr2_-_200329810
|
0.075
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr12_-_117742856
|
0.074
|
NM_001204213
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr21_-_45079327
|
0.071
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr8_-_103137134
|
0.070
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr8_-_103136486
|
0.070
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr8_-_102803438
|
0.070
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr12_+_13349601
|
0.069
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr2_-_27718098
|
0.068
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr16_-_28074776
|
0.068
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr2_-_218808705
|
0.065
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr22_+_38035683
|
0.065
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr12_-_117799410
|
0.064
|
NM_000620
NM_001204218
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr1_-_114354906
|
0.064
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr1_-_36851484
|
0.064
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr9_-_19786886
|
0.064
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr9_+_125133228
|
0.064
|
NM_000962
NM_080591
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr2_-_200322699
|
0.063
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr11_-_35441501
|
0.063
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr7_-_28220342
|
0.063
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr12_-_70083055
|
0.062
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr5_+_172068188
|
0.061
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr10_-_129924467
|
0.059
|
NM_001145966
NM_002417
|
MKI67
|
antigen identified by monoclonal antibody Ki-67
|
|
chr21_+_30452872
|
0.059
|
NM_020152
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chrX_+_68048792
|
0.058
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr2_+_30454396
|
0.058
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr7_+_105603656
|
0.058
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr8_+_27179918
|
0.057
|
NM_004103
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr8_+_27168953
|
0.057
|
NM_173174
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr8_+_27183062
|
0.057
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr3_-_98620452
|
0.056
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr7_+_18548899
|
0.055
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr14_-_95786095
|
0.055
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr7_+_18126543
|
0.054
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr8_+_27182995
|
0.054
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr20_+_33464327
|
0.054
|
NM_001076552
NM_018677
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr5_-_149682447
|
0.053
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr12_-_58160823
|
0.053
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr20_+_33462765
|
0.051
|
NM_001242393
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr12_-_118406027
|
0.051
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr17_+_30593194
|
0.050
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr18_+_47088400
|
0.050
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr12_-_42538432
|
0.048
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr5_-_151066492
|
0.047
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr22_+_40573879
|
0.047
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr2_+_7057471
|
0.047
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr22_+_40440664
|
0.047
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_-_208417635
|
0.046
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr17_+_4981721
|
0.046
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr16_+_77822400
|
0.046
|
NM_020927
|
VAT1L
|
vesicle amine transport protein 1 homolog (T. californica)-like
|
|
chr18_-_57364643
|
0.045
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr16_-_84651639
|
0.045
|
NM_021149
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr15_-_99789585
|
0.044
|
NM_001040655
NM_001040656
NM_001040657
NM_001040658
NM_001040659
NM_001040660
NM_022905
|
TTC23
|
tetratricopeptide repeat domain 23
|
|
chr12_+_49372235
|
0.044
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr15_+_71433787
|
0.044
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr17_-_1928177
|
0.044
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr16_-_27899477
|
0.044
|
NM_144675
|
GSG1L
|
GSG1-like
|
|
chr18_+_55102777
|
0.044
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr10_-_14372807
|
0.043
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr12_+_107712151
|
0.043
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr5_-_16617085
|
0.043
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr12_-_123214882
|
0.042
|
NM_032554
|
HCAR1
|
hydroxycarboxylic acid receptor 1
|
|
chr4_-_76928601
|
0.042
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr1_+_163038934
|
0.040
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr8_+_58906968
|
0.040
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr2_-_206950895
|
0.040
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr11_+_94501503
|
0.040
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr11_+_33563772
|
0.039
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr11_-_8615412
|
0.039
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr1_+_163038564
|
0.039
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr11_-_2160199
|
0.039
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr2_+_33701320
|
0.039
|
NM_001139488
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chrX_-_112066353
|
0.039
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr11_-_2162340
|
0.038
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr10_-_75634219
|
0.038
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr1_+_163038395
|
0.038
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chrX_+_106163605
|
0.037
|
NM_020384
|
CLDN2
|
claudin 2
|
|
chr11_+_15133969
|
0.037
|
NM_001031853
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr3_+_154798078
|
0.037
|
NM_007289
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_-_201476248
|
0.037
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr7_-_98467663
|
0.037
|
NM_001134450
NM_001134451
NM_152913
|
TMEM130
|
transmembrane protein 130
|
|
chr3_+_14166439
|
0.037
|
NM_024334
|
TMEM43
|
transmembrane protein 43
|
|
chr3_+_154797910
|
0.037
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_+_154797435
|
0.036
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr11_-_62783281
|
0.036
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chrX_-_71351750
|
0.036
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr11_+_15136483
|
0.036
|
NM_001042536
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr11_-_2170792
|
0.036
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr2_-_216878314
|
0.036
|
NM_018000
|
MREG
|
melanoregulin
|
|
chr2_+_9346876
|
0.036
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chrX_+_106143393
|
0.036
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr1_-_201475966
|
0.035
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr1_-_41131323
|
0.035
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr12_+_107974633
|
0.035
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr2_+_33738941
|
0.035
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr3_-_170626425
|
0.035
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr3_+_101498028
|
0.035
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr3_+_154797644
|
0.035
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr2_+_113033146
|
0.034
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr3_+_101498285
|
0.034
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr1_-_201465700
|
0.034
|
NM_001193572
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr19_-_3606801
|
0.034
|
NM_001060
NM_201636
|
TBXA2R
|
thromboxane A2 receptor
|
|
chr17_-_43394326
|
0.034
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr14_-_73925279
|
0.033
|
NM_001005743
NM_001005744
NM_001005745
NM_003744
|
NUMB
|
numb homolog (Drosophila)
|
|
chr11_+_119076974
|
0.033
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr2_+_33661392
|
0.033
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr1_+_44173217
|
0.033
|
NM_006279
NM_174963
NM_174964
NM_174965
NM_174966
NM_174967
NM_174968
NM_174969
NM_174970
NM_174971
|
ST3GAL3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3
|
|
chr1_+_163041694
|
0.032
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr16_-_17564737
|
0.032
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr5_+_71403040
|
0.032
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chrX_+_106161589
|
0.031
|
NM_001171095
|
CLDN2
|
claudin 2
|
|
chr10_-_98119058
|
0.031
|
NM_001040102
NM_001040103
NM_033207
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein
|
|
chr3_-_133614421
|
0.031
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr18_+_34409058
|
0.031
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr5_-_16509106
|
0.031
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr15_+_38544966
|
0.030
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr2_+_27805892
|
0.030
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr3_-_9291251
|
0.030
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr19_-_38146288
|
0.030
|
NM_014898
|
ZFP30
|
zinc finger protein 30 homolog (mouse)
|
|
chr17_+_64960754
|
0.030
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr5_-_149535420
|
0.030
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr7_+_50344255
|
0.029
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr17_-_79885544
|
0.029
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chrX_-_54384436
|
0.029
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr15_-_73661604
|
0.029
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr3_-_116164363
|
0.029
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr13_-_78492906
|
0.029
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr4_-_120548329
|
0.028
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr1_-_202612580
|
0.028
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr10_+_43572500
|
0.028
|
NM_020630
NM_020975
|
RET
|
ret proto-oncogene
|
|
chr12_+_54332575
|
0.028
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr11_+_66886735
|
0.028
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr22_-_31536339
|
0.027
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr15_-_34629941
|
0.027
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr3_+_57994087
|
0.027
|
NM_001164317
NM_001164318
NM_001164319
NM_001457
|
FLNB
|
filamin B, beta
|
|
chr11_+_122526339
|
0.027
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr15_-_70390229
|
0.027
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr15_-_40633122
|
0.027
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr21_-_40033617
|
0.027
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chrX_-_153095996
|
0.027
|
NM_032512
|
PDZD4
|
PDZ domain containing 4
|
|
chr4_+_184020343
|
0.026
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr13_-_33859835
|
0.026
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr15_-_34630203
|
0.026
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr5_-_11903964
|
0.026
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr2_+_27309610
|
0.026
|
NM_000221
NM_006488
|
KHK
|
ketohexokinase (fructokinase)
|
|
chr5_-_126366439
|
0.026
|
NM_178450
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr12_-_123594974
|
0.026
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr3_+_187871473
|
0.026
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr22_+_23412364
|
0.025
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr1_+_119911395
|
0.025
|
NM_001005783
NM_016527
|
HAO2
|
hydroxyacid oxidase 2 (long chain)
|
|
chr2_+_98962617
|
0.025
|
NM_001079878
NM_001298
|
CNGA3
|
cyclic nucleotide gated channel alpha 3
|
|
chrX_-_63425484
|
0.025
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr1_-_151804225
|
0.025
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr10_+_99400441
|
0.024
|
NM_018425
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha
|
|
chr20_-_43977023
|
0.024
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chrX_+_38660653
|
0.024
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr1_-_202679415
|
0.024
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr22_-_39240016
|
0.024
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr7_+_138916230
|
0.024
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr3_+_39851028
|
0.024
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr1_+_20208837
|
0.024
|
NM_015207
|
OTUD3
|
OTU domain containing 3
|
|
chr21_-_39870303
|
0.024
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr16_+_70613797
|
0.024
|
NM_001172771
NM_152456
|
IL34
|
interleukin 34
|
|
chr13_-_33760215
|
0.024
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr22_-_36784055
|
0.023
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr11_+_60681370
|
0.023
|
NM_024092
|
TMEM109
|
transmembrane protein 109
|
|
chr9_+_27109138
|
0.023
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr1_-_231560789
|
0.023
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr20_+_30697291
|
0.023
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr1_-_37499708
|
0.023
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr2_+_70142152
|
0.023
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr1_+_10270680
|
0.023
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr17_+_53342320
|
0.023
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr18_+_48405426
|
0.023
|
NM_001168335
NM_002396
|
ME2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial
|
|
chr22_-_45405429
|
0.023
|
NM_138415
|
PHF21B
|
PHD finger protein 21B
|
|
chr7_+_18535351
|
0.022
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr1_-_154832187
|
0.022
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr16_+_70657894
|
0.022
|
NM_001172772
|
IL34
|
interleukin 34
|
|
chr12_-_123451031
|
0.022
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|