|
chr7_+_100770378
|
4.941
|
NM_000602
NM_001165413
|
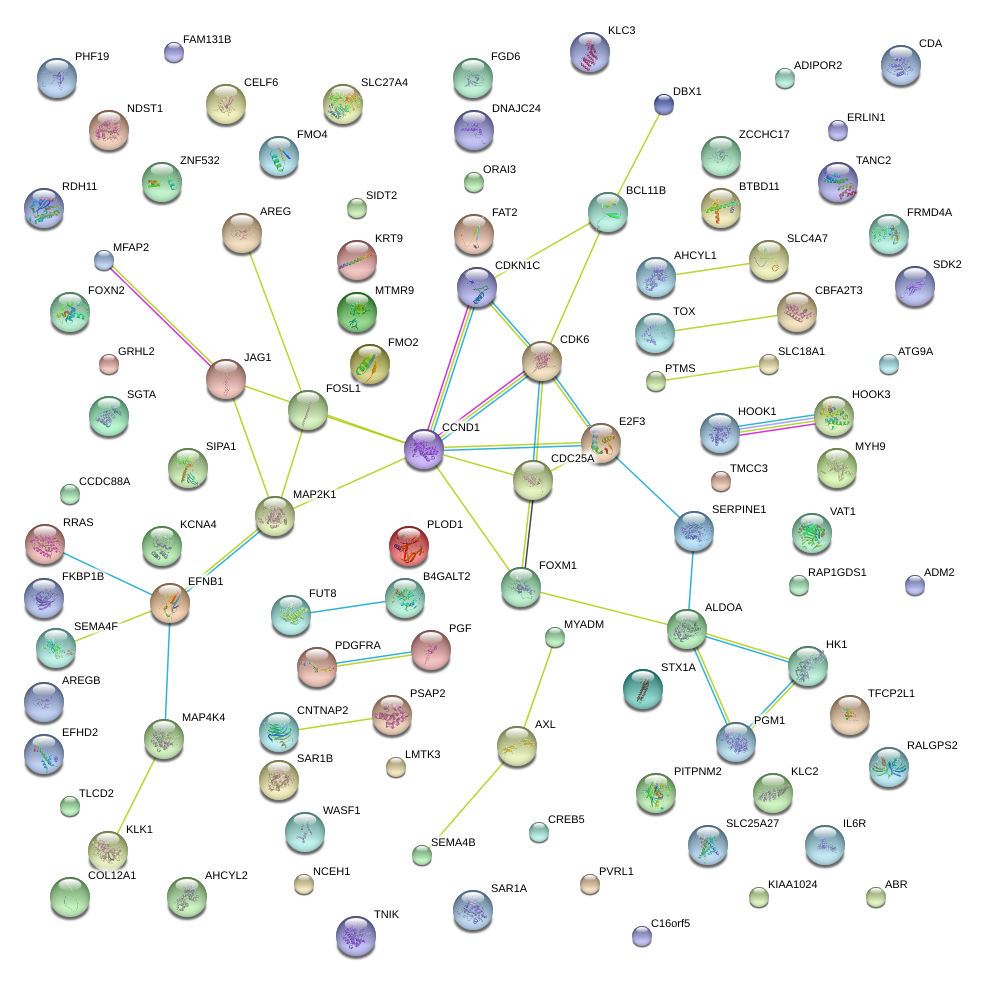
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr19_+_54371125
|
4.698
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr12_+_107712151
|
3.680
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr19_-_50143350
|
3.621
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr19_+_41725098
|
3.516
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr14_-_75422466
|
3.301
|
NM_001207012
NM_002632
|
PGF
|
placental growth factor
|
|
chr12_-_95044205
|
3.247
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr15_+_90728118
|
3.209
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr12_+_107974633
|
3.179
|
NM_001017523
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr19_+_54369610
|
3.146
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_-_49016418
|
2.970
|
NM_001080434
|
LMTK3
|
lemur tyrosine kinase 3
|
|
chr2_+_48541767
|
2.491
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr19_+_54372659
|
2.487
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_-_2783332
|
2.485
|
NM_003021
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha
|
|
chr6_+_20402040
|
2.442
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr11_-_65667860
|
2.409
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr7_+_145813452
|
2.298
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr2_+_24272583
|
2.248
|
NM_004116
NM_054033
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa
|
|
chr17_-_41174431
|
2.194
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr7_-_73133917
|
2.174
|
NM_001165903
NM_004603
|
STX1A
|
syntaxin 1A (brain)
|
|
chr11_-_2906960
|
2.166
|
NM_000076
NM_001122630
NM_001122631
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr15_+_90744506
|
2.150
|
NM_198925
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr12_+_1800188
|
2.089
|
NM_024551
|
ADIPOR2
|
adiponectin receptor 2
|
|
chr16_-_4588738
|
2.053
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr16_-_4588379
|
1.916
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr17_-_71640226
|
1.909
|
NM_001144952
|
SDK2
|
sidekick cell adhesion molecule 2
|
|
chrX_+_68048792
|
1.880
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr10_-_14372807
|
1.796
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr12_-_95611023
|
1.769
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr15_+_66679107
|
1.759
|
NM_002755
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr2_-_220094354
|
1.748
|
NM_001077198
NM_024085
|
ATG9A
|
ATG9 autophagy related 9 homolog A (S. cerevisiae)
|
|
chr3_-_27498244
|
1.745
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr18_+_56530060
|
1.714
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr1_+_178694273
|
1.712
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr4_+_99182520
|
1.626
|
NM_001100426
NM_001100427
NM_001100428
NM_001100429
NM_001100430
NM_021159
|
RAP1GDS1
|
RAP1, GTP-GDP dissociation stimulator 1
|
|
chr2_+_102314162
|
1.615
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr11_+_117049838
|
1.579
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr6_+_46620631
|
1.570
|
NM_001204051
NM_001204052
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr17_+_61086897
|
1.541
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr6_-_75915566
|
1.518
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr15_-_72598655
|
1.496
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr8_+_42752019
|
1.480
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr16_+_30960377
|
1.458
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr10_-_101945686
|
1.450
|
NM_006459
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr22_+_51113069
|
1.443
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr10_-_71930185
|
1.431
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr10_-_101945787
|
1.418
|
NM_001100626
|
ERLIN1
|
ER lipid raft associated 1
|
|
chr11_+_31391371
|
1.413
|
NM_181706
|
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24
|
|
chr4_+_75480628
|
1.411
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr2_-_122042767
|
1.377
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr8_-_60031545
|
1.371
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr8_+_11141983
|
1.366
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr4_+_75310852
|
1.341
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr9_+_131102786
|
1.335
|
NM_005094
|
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4
|
|
chr5_-_150948504
|
1.332
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr15_+_79724857
|
1.312
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr11_+_65405538
|
1.309
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr14_-_99737821
|
1.281
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr5_+_149887673
|
1.273
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr7_+_129015483
|
1.257
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr7_-_92463211
|
1.247
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr16_+_30064410
|
1.246
|
NM_000034
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr2_-_55646946
|
1.244
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr12_-_2986299
|
1.232
|
NM_001243088
NM_001243089
NM_021953
NM_202002
NM_202003
|
FOXM1
|
forkhead box M1
|
|
chr7_-_143059696
|
1.232
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr7_-_143059144
|
1.229
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr3_-_172428773
|
1.225
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr4_+_55095263
|
1.224
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr1_+_64088886
|
1.199
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr14_+_65877309
|
1.199
|
NM_004480
NM_178155
NM_178156
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr1_+_64059479
|
1.199
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr1_+_11994710
|
1.198
|
NM_000302
|
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1
|
|
chr6_-_110501164
|
1.197
|
NM_001024934
NM_001024935
NM_001024936
NM_003931
|
WASF1
|
WAS protein family, member 1
|
|
chr11_+_69455837
|
1.192
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr1_+_44446176
|
1.184
|
NM_001005417
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr8_+_102504661
|
1.182
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_+_31769826
|
1.166
|
NM_016505
|
ZCCHC17
|
zinc finger, CCHC domain containing 17
|
|
chr1_-_17307107
|
1.166
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr11_+_66024746
|
1.158
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr10_+_71029755
|
1.155
|
NM_033497
NM_033498
NM_033500
|
HK1
|
hexokinase 1
|
|
chr12_-_123594974
|
1.148
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr16_-_89007605
|
1.144
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr20_-_10654429
|
1.144
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr7_+_129007942
|
1.142
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr22_-_36784055
|
1.136
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr2_+_74881353
|
1.123
|
NM_004263
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F
|
|
chr7_+_28338939
|
1.116
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_-_123639566
|
1.094
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr1_-_17308072
|
1.091
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr3_-_48229800
|
1.080
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr17_-_1613661
|
1.050
|
NM_001164407
|
TLCD2
|
TLC domain containing 2
|
|
chr7_-_92465929
|
1.043
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr1_+_20915440
|
1.043
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr14_-_68162419
|
1.040
|
NM_001252650
NM_016026
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis)
|
|
chr17_-_1012257
|
1.032
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr22_+_50919970
|
1.007
|
NM_024866
NM_001253845
|
ADM2
|
adrenomedullin 2
|
|
chr1_+_15736387
|
1.006
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr11_-_119599253
|
1.006
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr1_+_171154387
|
1.003
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr1_+_154377668
|
1.002
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr3_+_25469749
|
0.996
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr3_+_38206968
|
0.978
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr7_+_28725597
|
0.970
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr2_+_113875469
|
0.968
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr17_+_70117154
|
0.962
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr19_-_14889347
|
0.962
|
NM_013447
NM_152916
NM_152917
NM_152918
NM_152919
NM_152920
NM_152921
|
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2
|
|
chr12_-_109125289
|
0.956
|
NM_014325
|
CORO1C
|
coronin, actin binding protein, 1C
|
|
chr1_+_165796652
|
0.956
|
NM_012474
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr7_+_128864740
|
0.955
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr17_+_1646290
|
0.952
|
NM_001165920
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr3_-_113465101
|
0.950
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr4_+_144257982
|
0.943
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr1_-_21044202
|
0.937
|
NM_001122819
NM_020816
|
KIF17
|
kinesin family member 17
|
|
chr16_+_81069316
|
0.937
|
NM_015251
|
ATMIN
|
ATM interactor
|
|
chr12_+_49208214
|
0.929
|
NM_001206917
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr1_+_64058891
|
0.919
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr2_+_232651059
|
0.916
|
NM_022730
|
COPS7B
|
COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis)
|
|
chr17_+_1646129
|
0.915
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr1_+_44445548
|
0.901
|
NM_030587
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr5_+_175298492
|
0.899
|
NM_001008220
|
CPLX2
|
complexin 2
|
|
chr3_+_171757413
|
0.892
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr5_+_172261220
|
0.890
|
NM_001031711
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1
|
|
chr1_+_231298673
|
0.886
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr3_+_135741576
|
0.884
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr5_+_150399980
|
0.882
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr9_-_139440237
|
0.881
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr1_+_201708940
|
0.877
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr1_+_44444848
|
0.870
|
NM_003780
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2
|
|
chr9_-_4741267
|
0.865
|
NM_001199852
NM_016282
|
AK3
|
adenylate kinase 3
|
|
chr11_-_9025531
|
0.864
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr2_+_113885137
|
0.857
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr19_-_2456954
|
0.854
|
NM_032737
|
LMNB2
|
lamin B2
|
|
chr3_+_171758289
|
0.851
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr5_+_175223519
|
0.847
|
NM_006650
|
CPLX2
|
complexin 2
|
|
chr6_-_30655671
|
0.846
|
NM_133471
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr18_-_28622624
|
0.838
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr12_+_49209300
|
0.833
|
NM_001206916
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr7_+_28452143
|
0.833
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_+_88718287
|
0.827
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr14_+_23299083
|
0.824
|
NM_178336
NM_180982
NM_181304
NM_181305
NM_181306
NM_181307
|
MRPL52
|
mitochondrial ribosomal protein L52
|
|
chr12_+_107168336
|
0.821
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr19_+_7911385
|
0.820
|
NM_001159944
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr2_-_167005641
|
0.817
|
NM_001202435
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr7_+_28475233
|
0.814
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_+_105036783
|
0.813
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr17_-_8066254
|
0.806
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr5_-_149535420
|
0.805
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr9_-_4741085
|
0.803
|
NM_001199854
|
AK3
|
adenylate kinase 3
|
|
chr2_+_95691428
|
0.791
|
NM_002371
NM_022438
NM_022439
NM_022440
|
MAL
|
mal, T-cell differentiation protein
|
|
chr12_+_49212253
|
0.789
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr6_-_30655078
|
0.786
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr17_-_16472463
|
0.777
|
NM_020653
|
ZNF287
|
zinc finger protein 287
|
|
chr9_+_112887780
|
0.775
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr8_+_28174648
|
0.770
|
NM_006228
|
PNOC
|
prepronociceptin
|
|
chr9_-_4741798
|
0.760
|
NM_001199855
|
AK3
|
adenylate kinase 3
|
|
chr2_-_47143250
|
0.756
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr9_-_4742042
|
0.756
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr1_+_66258855
|
0.753
|
NM_002600
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr3_+_50273645
|
0.752
|
NM_002070
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr5_-_132299263
|
0.750
|
NM_014423
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr9_-_4726226
|
0.748
|
NM_001199856
|
AK3
|
adenylate kinase 3
|
|
chr2_+_45169036
|
0.742
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chr1_+_201617449
|
0.740
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr2_-_31440378
|
0.740
|
NM_001145122
|
CAPN14
|
calpain 14
|
|
chr14_-_54955714
|
0.739
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr6_+_150263120
|
0.738
|
NM_025217
|
ULBP2
|
UL16 binding protein 2
|
|
chr11_+_46354454
|
0.738
|
NM_201532
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr9_+_124329398
|
0.736
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr9_+_118916069
|
0.734
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr1_+_160175114
|
0.732
|
NM_003768
|
PEA15
|
phosphoprotein enriched in astrocytes 15
|
|
chr3_+_47324329
|
0.726
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr11_+_46368881
|
0.717
|
NM_001199266
NM_001199267
NM_001199268
NM_003646
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr10_+_11784355
|
0.712
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr11_+_18417812
|
0.709
|
NM_001165414
|
LDHA
|
lactate dehydrogenase A
|
|
chr3_+_50284325
|
0.709
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr11_-_66488712
|
0.705
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr22_-_36424397
|
0.704
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr2_+_220325533
|
0.704
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chr17_-_31620005
|
0.702
|
NM_183377
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr11_+_65407591
|
0.702
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr3_-_56835829
|
0.696
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chrX_-_63425484
|
0.693
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr10_-_15210610
|
0.682
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chrX_-_18372777
|
0.680
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr2_-_27485774
|
0.680
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr7_+_138916230
|
0.677
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr9_+_112810877
|
0.676
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr10_+_99205913
|
0.674
|
NM_032327
NM_198043
NM_198044
NM_198045
NM_198046
|
ZDHHC16
|
zinc finger, DHHC-type containing 16
|
|
chr17_+_4901199
|
0.670
|
NM_006612
|
KIF1C
|
kinesin family member 1C
|
|
chr17_-_32483550
|
0.668
|
NM_001094
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr17_-_36762105
|
0.667
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr14_-_64194692
|
0.666
|
NM_030791
|
SGPP1
|
sphingosine-1-phosphate phosphatase 1
|
|
chr22_-_29977325
|
0.665
|
NM_003634
|
NIPSNAP1
|
nipsnap homolog 1 (C. elegans)
|
|
chr21_+_41029253
|
0.665
|
NM_006057
NM_033170
NM_033171
NM_033172
NM_033173
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5
|
|
chr18_+_55019720
|
0.658
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr2_-_47142950
|
0.657
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr11_-_118047336
|
0.654
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr17_+_43971655
|
0.653
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr18_-_48724031
|
0.649
|
NM_016626
|
MEX3C
|
mex-3 homolog C (C. elegans)
|
|
chr3_+_10206562
|
0.646
|
NM_001570
|
IRAK2
|
interleukin-1 receptor-associated kinase 2
|