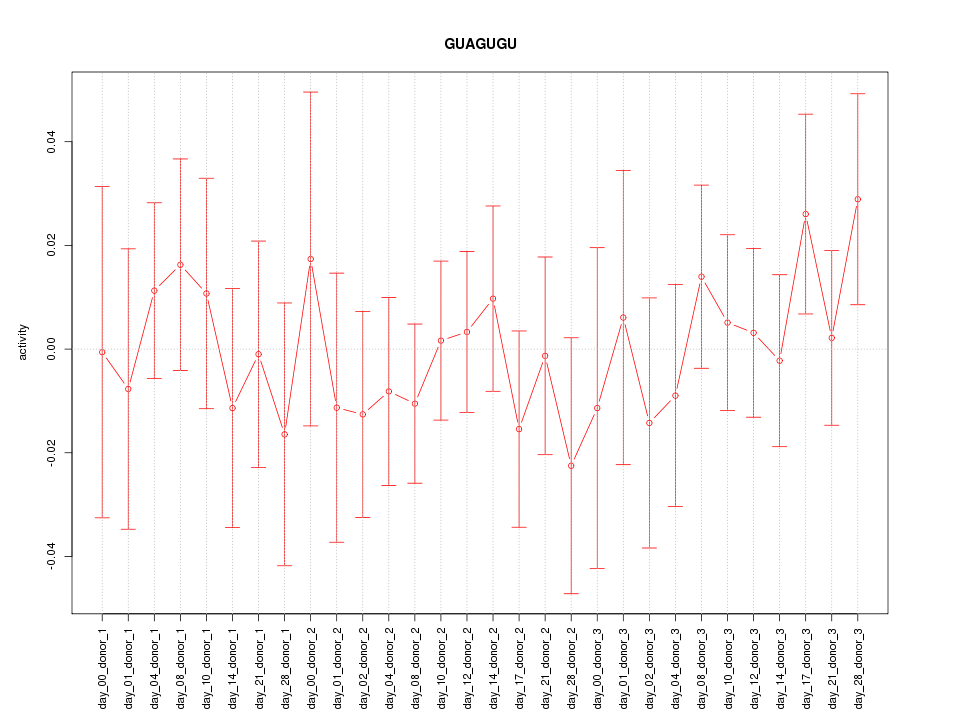
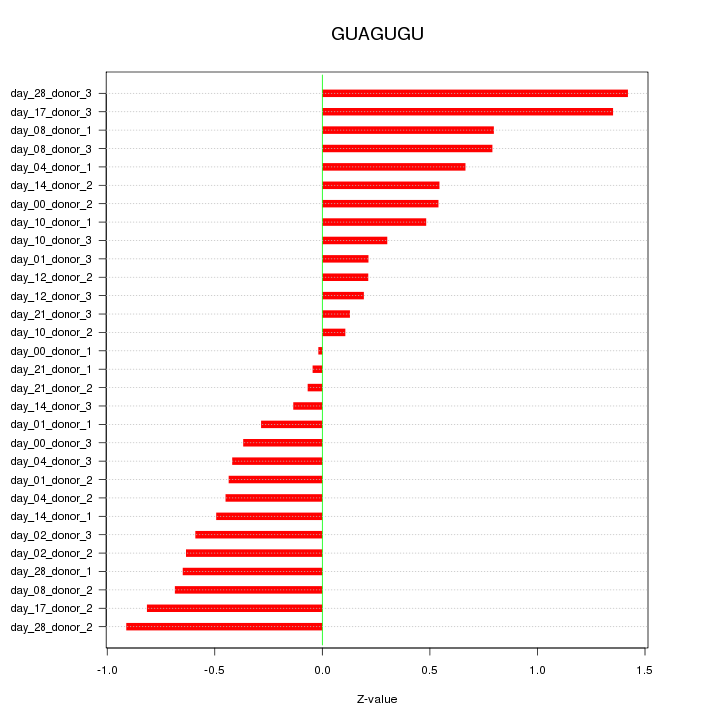
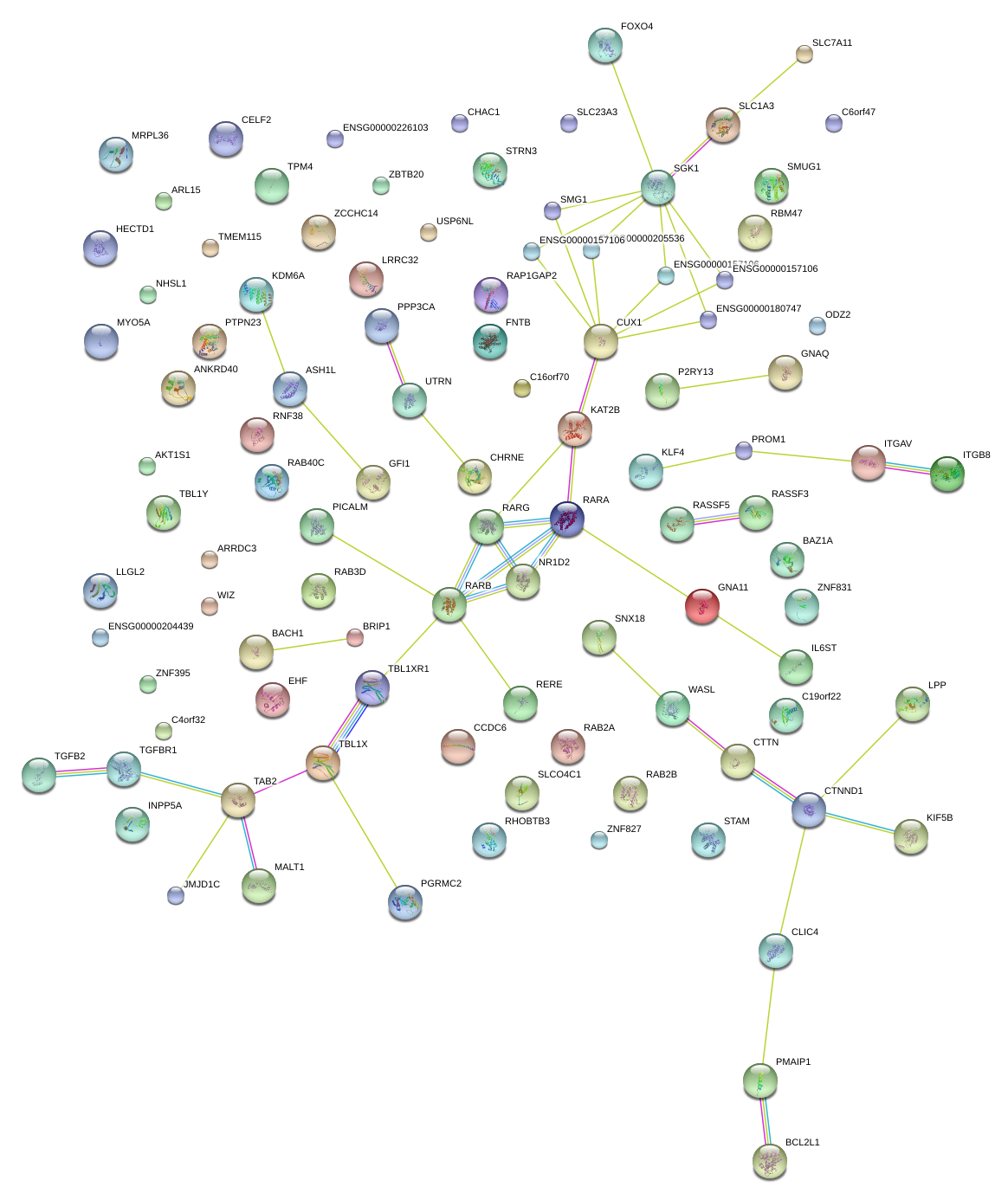
|
chr4_-_16077740
|
1.038
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr4_-_16085593
|
0.984
|
NM_001145847
NM_001145848
|
PROM1
|
prominin 1
|
|
chr11_-_76381787
|
0.691
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr11_-_76380894
|
0.681
|
NM_005512
|
LRRC32
|
leucine rich repeat containing 32
|
|
chrX_+_44732402
|
0.559
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr17_-_4806368
|
0.524
|
NM_000080
|
CHRNE
|
cholinergic receptor, nicotinic, epsilon
|
|
chr1_-_29450406
|
0.450
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr1_-_29449012
|
0.422
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr1_+_25071759
|
0.398
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr6_-_138820578
|
0.386
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr11_+_34654010
|
0.367
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr4_-_139163222
|
0.367
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr6_-_138893666
|
0.349
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr9_-_80646150
|
0.349
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr5_-_55290601
|
0.336
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr11_+_34642587
|
0.330
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr4_+_113066552
|
0.323
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr7_+_20370724
|
0.322
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr1_+_218518675
|
0.321
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr5_+_95066628
|
0.306
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr14_+_65453439
|
0.293
|
NM_002028
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr18_+_56338617
|
0.285
|
NM_006785
NM_173844
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr19_+_45596406
|
0.279
|
NM_019121
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37
|
|
chr10_-_11574273
|
0.276
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr3_-_151047309
|
0.270
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr5_+_166711842
|
0.268
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr19_+_16178170
|
0.264
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr12_+_65004191
|
0.254
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr4_-_40631846
|
0.251
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr1_-_155532322
|
0.244
|
NM_018489
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr14_-_31495524
|
0.239
|
NM_001083893
NM_014574
|
STRN3
|
striatin, calmodulin binding protein 3
|
|
chr10_+_5726781
|
0.238
|
NM_017782
|
FAM208B
|
family with sequence similarity 208, member B
|
|
chr4_-_146859568
|
0.235
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr6_+_144612872
|
0.234
|
NM_007124
|
UTRN
|
utrophin
|
|
chr11_-_85780899
|
0.227
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_54582588
|
0.223
|
NM_001243789
NM_001243790
NM_001243787
NM_001243788
NM_001243791
NM_014311
|
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1
|
|
chr9_-_110252045
|
0.218
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr19_+_16187047
|
0.218
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr7_-_123388711
|
0.216
|
NM_003941
|
WASL
|
Wiskott-Aldrich syndrome-like
|
|
chr3_+_187943192
|
0.215
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr19_-_11450254
|
0.214
|
NM_004283
|
RAB3D
|
RAB3D, member RAS oncogene family
|
|
chr5_-_90679115
|
0.210
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr3_+_20081523
|
0.199
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chr5_+_36608430
|
0.197
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_-_114866090
|
0.196
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_+_101867401
|
0.196
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr7_+_101460881
|
0.195
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr4_-_40517913
|
0.193
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr17_-_48785016
|
0.192
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr16_-_87525317
|
0.190
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr2_+_198380294
|
0.188
|
NM_199482
|
MOB4
|
MOB family member 4, phocein
|
|
chr15_-_52821004
|
0.184
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr6_+_149639435
|
0.182
|
NM_015093
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2
|
|
chr21_+_30671736
|
0.182
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr14_-_31676680
|
0.179
|
NM_015382
|
HECTD1
|
HECT domain containing 1
|
|
chr3_-_50396930
|
0.178
|
NM_007024
|
TMEM115
|
transmembrane protein 115
|
|
chr5_-_101631965
|
0.176
|
NM_180991
|
SLCO4C1
|
solute carrier organic anion transporter family, member 4C1
|
|
chr12_-_53614121
|
0.173
|
NM_001042728
NM_001243732
|
RARG
|
retinoic acid receptor, gamma
|
|
chr18_+_57567191
|
0.172
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr4_-_129209983
|
0.172
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr3_+_47422471
|
0.171
|
NM_015466
|
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23
|
|
chr3_+_187930720
|
0.171
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr6_-_134498973
|
0.171
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134639195
|
0.170
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_+_17686123
|
0.168
|
NM_003473
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1
|
|
chr20_+_57766074
|
0.165
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr1_-_8483701
|
0.162
|
NM_001042682
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr19_-_50380451
|
0.159
|
NM_032375
NM_001098633
NM_001098632
|
AKT1S1
|
AKT1 substrate 1 (proline-rich)
|
|
chr12_-_53625834
|
0.159
|
NM_000966
NM_001243730
NM_001243731
|
RARG
|
retinoic acid receptor, gamma
|
|
chr10_-_61666236
|
0.156
|
NM_005436
|
CCDC6
|
coiled-coil domain containing 6
|
|
chr1_-_92952429
|
0.156
|
NM_005263
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr20_-_30310597
|
0.154
|
NM_001191
NM_138578
|
BCL2L1
|
BCL2-like 1
|
|
chr16_+_67143783
|
0.154
|
NM_025187
|
C16orf70
|
chromosome 16 open reading frame 70
|
|
chr15_+_41245635
|
0.153
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr8_+_61429390
|
0.152
|
NM_001242644
NM_002865
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr6_-_134496006
|
0.152
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chrX_+_70315637
|
0.150
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr16_+_640172
|
0.147
|
NM_021168
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr2_+_187464931
|
0.146
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr16_+_639356
|
0.144
|
NM_001172663
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr11_-_85780105
|
0.144
|
NM_001008660
NM_001206946
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_-_8877379
|
0.142
|
NM_001042681
NM_012102
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr3_+_23987514
|
0.139
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr14_-_35344852
|
0.139
|
NM_013448
NM_182648
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr17_+_2699731
|
0.138
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr1_-_92949355
|
0.137
|
NM_001127215
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr16_-_18937722
|
0.137
|
NM_015092
|
SMG1
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr11_+_57529233
|
0.136
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr9_-_36400212
|
0.136
|
NM_022781
NM_194329
|
RNF38
|
ring finger protein 38
|
|
chrX_+_9431314
|
0.134
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr3_-_114478117
|
0.133
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr8_-_28243941
|
0.133
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr6_-_31628509
|
0.133
|
NM_021184
|
C6orf47
|
chromosome 6 open reading frame 47
|
|
chr4_-_102268626
|
0.133
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr10_+_134351272
|
0.133
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr3_+_23986735
|
0.132
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr5_-_53606387
|
0.129
|
NM_019087
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr10_+_11047253
|
0.129
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_-_32345337
|
0.128
|
NM_004521
|
KIF5B
|
kinesin family member 5B
|
|
chr6_-_134497069
|
0.127
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_+_53813494
|
0.126
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chr10_-_65028825
|
0.126
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr17_+_73521744
|
0.124
|
NM_001015002
NM_001031803
NM_004524
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr11_+_70244588
|
0.120
|
NM_001184740
NM_005231
NM_138565
|
CTTN
|
cortactin
|
|
chr19_-_15560761
|
0.119
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr9_-_36401149
|
0.118
|
NM_194330
|
RNF38
|
ring finger protein 38
|
|
chr18_+_67956136
|
0.118
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr1_+_57110745
|
0.117
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr10_-_61900659
|
0.116
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr6_-_88411984
|
0.115
|
NM_018064
|
AKIRIN2
|
akirin 2
|
|
chr20_-_3996035
|
0.114
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr8_-_17579729
|
0.114
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr17_+_7210317
|
0.112
|
NM_001143760
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr17_+_27717942
|
0.111
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|
|
chr16_-_70557430
|
0.111
|
NM_001195139
NM_015386
|
COG4
|
component of oligomeric golgi complex 4
|
|
chr9_-_36400817
|
0.111
|
NM_194328
NM_194332
|
RNF38
|
ring finger protein 38
|
|
chrX_-_24045302
|
0.110
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chrX_-_77914824
|
0.109
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr8_-_17555158
|
0.109
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr10_-_27529263
|
0.108
|
NM_145698
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chrX_+_16964730
|
0.107
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr10_-_27531041
|
0.107
|
NM_001042473
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr3_+_187871473
|
0.105
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_-_91487769
|
0.105
|
NM_032186
NM_201269
|
ZNF644
|
zinc finger protein 644
|
|
chr8_-_17580024
|
0.104
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr18_-_33647336
|
0.104
|
NM_018170
|
RPRD1A
|
regulation of nuclear pre-mRNA domain containing 1A
|
|
chr21_+_30671217
|
0.103
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr16_+_640056
|
0.103
|
NM_001172665
NM_001172666
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr2_+_198381021
|
0.102
|
NM_001204094
|
MOB4
|
MOB family member 4, phocein
|
|
chr2_+_148602569
|
0.101
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr2_+_198380752
|
0.101
|
NM_001100819
NM_015387
|
MOB4
|
MOB family member 4, phocein
|
|
chr10_-_81205091
|
0.101
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr7_-_112726143
|
0.099
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr5_+_42549657
|
0.099
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr2_+_187454724
|
0.099
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chrX_-_41782230
|
0.097
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr1_+_11072678
|
0.097
|
NM_007375
|
TARDBP
|
TAR DNA binding protein
|
|
chrX_+_122993536
|
0.097
|
NM_001204401
NM_001167
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr19_+_56187990
|
0.097
|
NM_001130071
|
EPN1
|
epsin 1
|
|
chr17_+_7211693
|
0.096
|
NM_001143762
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr16_+_28875181
|
0.094
|
NM_001145812
NM_015503
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr3_-_114790221
|
0.094
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_42550181
|
0.093
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr16_+_639668
|
0.092
|
NM_001172664
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr15_+_38544966
|
0.092
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr11_+_13299260
|
0.091
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr8_-_17658385
|
0.090
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_+_42565562
|
0.090
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr5_+_42548314
|
0.089
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr11_-_8680382
|
0.088
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr2_-_39664129
|
0.088
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr7_-_112727832
|
0.088
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr14_-_35182944
|
0.087
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr5_+_42565963
|
0.087
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chrX_+_9432980
|
0.087
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr10_-_62149487
|
0.086
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr5_+_42548144
|
0.086
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr11_+_102188180
|
0.086
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr17_+_44668034
|
0.084
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr6_-_159466019
|
0.083
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr5_+_42424553
|
0.083
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr10_-_32217762
|
0.082
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chrX_-_109683457
|
0.081
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr4_+_15004297
|
0.081
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr18_+_42260774
|
0.081
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chrX_-_102942968
|
0.081
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr5_+_112073555
|
0.081
|
NM_000038
NM_001127510
|
APC
|
adenomatous polyposis coli
|
|
chr16_+_28875077
|
0.081
|
NM_001145795
NM_001145796
NM_001145797
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr2_+_74273449
|
0.081
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr2_-_99347588
|
0.081
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr5_+_42425093
|
0.079
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr9_-_115095933
|
0.079
|
NM_001163788
NM_001163790
NM_001244896
NM_001244897
NM_005156
|
PTBP3
|
polypyrimidine tract binding protein 3
|
|
chr6_-_132834165
|
0.079
|
NM_003569
|
STX7
|
syntaxin 7
|
|
chr3_-_114102488
|
0.079
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_+_42423811
|
0.078
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr3_+_238274
|
0.077
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr22_-_36784055
|
0.077
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr12_-_44200047
|
0.076
|
NM_001242397
NM_002822
|
TWF1
|
twinfilin, actin-binding protein, homolog 1 (Drosophila)
|
|
chr5_-_142815076
|
0.076
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr6_+_34206567
|
0.074
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr17_-_47439326
|
0.074
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr19_-_22379752
|
0.073
|
NM_001001411
|
ZNF676
|
zinc finger protein 676
|
|
chr3_-_114343791
|
0.071
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_7210841
|
0.071
|
NM_001970
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr2_-_86948244
|
0.071
|
NM_001198954
|
RNF103-CHMP3
|
RNF103-CHMP3 readthrough
|
|
chr17_+_7211260
|
0.071
|
NM_001143761
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr17_-_62340652
|
0.070
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr14_-_35183722
|
0.068
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr3_-_114343052
|
0.068
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_+_224301790
|
0.068
|
NM_001136115
NM_015176
|
FBXO28
|
F-box protein 28
|
|
chr5_+_31799030
|
0.067
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr6_-_33267128
|
0.066
|
NM_001243738
NM_004761
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr15_+_66161769
|
0.065
|
NM_001206836
NM_004663
|
RAB11A
|
RAB11A, member RAS oncogene family
|
|
chr19_-_6279927
|
0.065
|
NM_005934
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chr2_-_61765394
|
0.063
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr21_+_18885104
|
0.063
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr1_+_114493766
|
0.063
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr8_+_81398416
|
0.063
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr5_+_112043188
|
0.063
|
NM_001127511
|
APC
|
adenomatous polyposis coli
|
|
chr8_-_116681103
|
0.062
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|