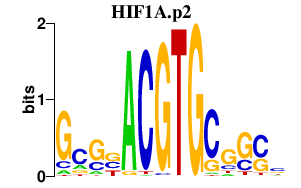
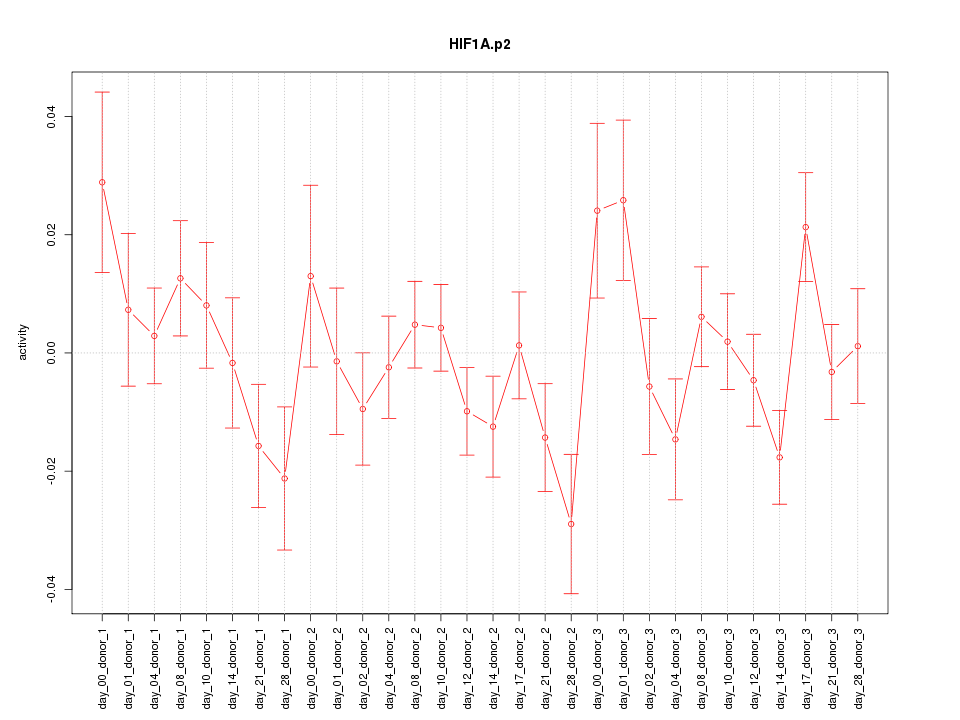
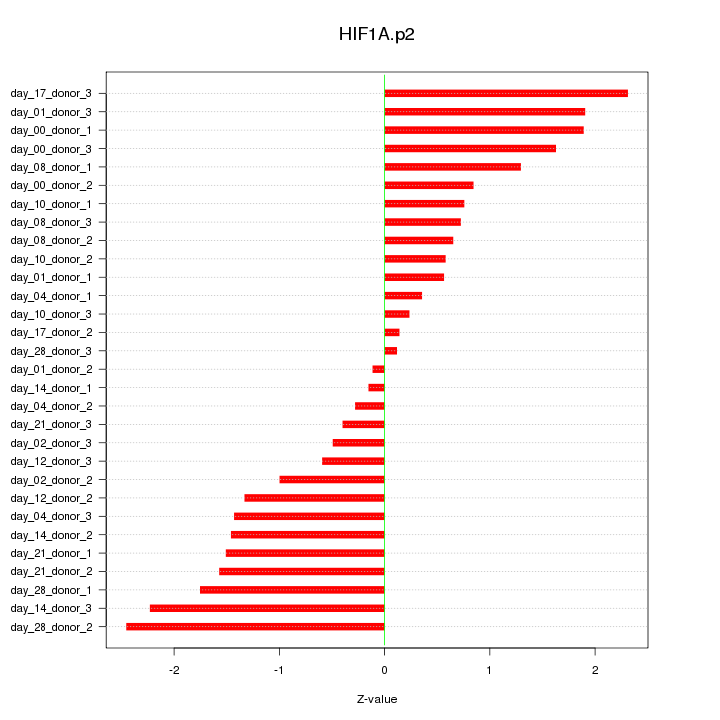
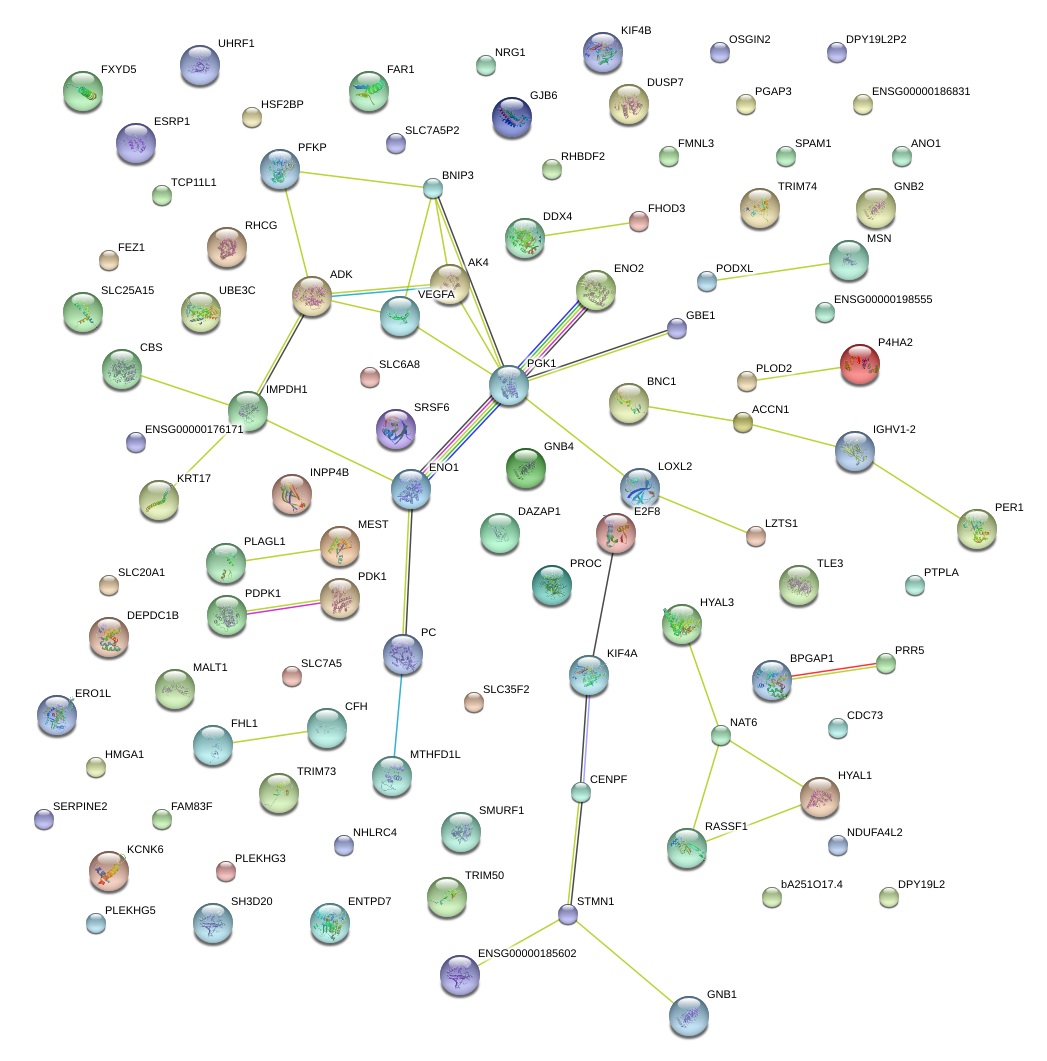
|
chr8_-_23261626
|
3.138
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr8_-_23261632
|
2.801
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr13_-_20806533
|
2.684
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr1_+_193091283
|
2.631
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr8_-_23261675
|
2.560
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr6_+_43738757
|
2.523
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chrX_+_69509878
|
2.365
|
NM_012310
|
KIF4A
|
kinesin family member 4A
|
|
chr12_-_57634474
|
2.291
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr16_-_87903028
|
2.226
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr5_-_59995938
|
2.223
|
NM_001145208
NM_018369
|
DEPDC1B
|
DEP domain containing 1B
|
|
chr15_-_90039776
|
2.219
|
NM_016321
|
RHCG
|
Rh family, C glycoprotein
|
|
chr22_+_40390935
|
2.194
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chrX_+_135229536
|
2.092
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_-_102920665
|
2.051
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr5_+_154393259
|
2.030
|
NM_001099293
|
KIF4B
|
kinesin family member 4B
|
|
chr11_+_69924407
|
1.966
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr7_-_102920840
|
1.958
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr1_-_26233367
|
1.907
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr8_-_23261591
|
1.900
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr19_+_4909447
|
1.865
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr2_+_173420873
|
1.863
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr1_-_26232890
|
1.850
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr7_-_98741605
|
1.802
|
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr8_+_31497267
|
1.727
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr7_-_98741681
|
1.632
|
NM_001199847
NM_020429
NM_181349
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1
|
|
chr10_+_3110818
|
1.627
|
NM_001242339
|
PFKP
|
phosphofructokinase, platelet
|
|
chr3_-_81810618
|
1.583
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr13_+_41363754
|
1.583
|
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr3_-_50336662
|
1.536
|
NM_001200016
NM_012191
NM_001200030
NM_001200031
NM_001200032
NM_003549
|
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related)
hyaluronoglucosaminidase 3
|
|
chr2_+_173420794
|
1.533
|
|
PDK1
|
pyruvate dehydrogenase kinase, isozyme 1
|
|
chr3_-_145879222
|
1.513
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr21_-_44495894
|
1.510
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr7_+_27135672
|
1.503
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr3_-_81810914
|
1.501
|
NM_000158
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr15_-_83953465
|
1.496
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr7_+_156931663
|
1.479
|
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr11_-_568419
|
1.444
|
|
MIR210HG
|
MIR210 host gene (non-protein coding)
|
|
chr11_-_125366048
|
1.444
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr2_-_224903933
|
1.443
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr3_-_50336258
|
1.432
|
NM_001200018
NM_001200029
|
NAT6
HYAL3
|
N-acetyltransferase 6 (GCN5-related)
hyaluronoglucosaminidase 3
|
|
chr7_+_128095881
|
1.420
|
NM_001098786
NM_013332
|
HILPDA
|
hypoxia inducible lipid droplet-associated
|
|
chr7_+_130126035
|
1.412
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr20_+_42086533
|
1.405
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr6_+_34204690
|
1.358
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr11_-_66725790
|
1.356
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr6_+_34205029
|
1.347
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr3_-_81810816
|
1.324
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr1_+_65613211
|
1.302
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr1_-_26232643
|
1.291
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr3_-_50374795
|
1.270
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chrX_+_64887534
|
1.267
|
|
MSN
|
moesin
|
|
chr14_+_65171243
|
1.263
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chrX_+_64887510
|
1.263
|
NM_002444
|
MSN
|
moesin
|
|
chr5_-_131563481
|
1.240
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr6_+_151187310
|
1.234
|
NM_001242768
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr7_+_156931631
|
1.223
|
NM_014671
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr19_+_38810540
|
1.204
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr19_+_35646006
|
1.203
|
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr18_+_33877630
|
1.202
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr11_+_13690246
|
1.200
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr6_+_151187064
|
1.196
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr1_+_65613971
|
1.193
|
|
AK4
|
adenylate kinase 4
|
|
chr2_+_113403433
|
1.174
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr3_-_52090090
|
1.169
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr22_+_45148437
|
1.165
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr6_+_34204928
|
1.162
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr4_-_143767603
|
1.159
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chr6_+_34204663
|
1.154
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chrX_+_152954965
|
1.150
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr13_+_41363642
|
1.146
|
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr1_+_214776494
|
1.141
|
NM_016343
|
CENPF
|
centromere protein F, 350/400kDa (mitosin)
|
|
chr13_+_41363546
|
1.140
|
NM_014252
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15
|
|
chr14_-_53162198
|
1.123
|
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr10_-_17659365
|
1.117
|
NM_014241
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A
|
|
chr1_-_8939150
|
1.111
|
NM_001428
|
ENO1
|
enolase 1, (alpha)
|
|
chr3_-_145878936
|
1.111
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr3_-_179169180
|
1.107
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr21_-_45079327
|
1.100
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr17_-_43510279
|
1.090
|
NM_174919
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr10_-_133795311
|
1.066
|
NM_004052
|
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3
|
|
chr6_+_34205369
|
1.065
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr14_-_53162370
|
1.039
|
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr17_-_74497406
|
1.038
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chrX_+_77359665
|
1.028
|
NM_000291
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr14_-_53162418
|
1.024
|
NM_014584
|
ERO1L
|
ERO1-like (S. cerevisiae)
|
|
chr16_+_610393
|
1.009
|
NM_145270
|
C16orf11
|
chromosome 16 open reading frame 11
|
|
chr11_-_19263144
|
1.003
|
|
E2F8
|
E2F transcription factor 8
|
|
chr1_+_214776582
|
0.998
|
|
CENPF
|
centromere protein F, 350/400kDa (mitosin)
|
|
chr19_+_1407750
|
0.994
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr10_-_133795447
|
0.993
|
|
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3
|
|
chr11_+_33061560
|
0.987
|
NM_001145541
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr5_+_55033844
|
0.983
|
NM_001142549
NM_001166533
NM_024415
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr1_-_6557448
|
0.970
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr8_+_95653272
|
0.959
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr10_+_101419169
|
0.956
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr6_-_144385647
|
0.946
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr15_-_70390226
|
0.935
|
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr11_+_33061291
|
0.930
|
|
TCP11L1
|
t-complex 11 (mouse)-like 1
|
|
chr1_-_29450406
|
0.914
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr17_-_8055684
|
0.911
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr11_-_107729492
|
0.907
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr7_-_128045995
|
0.906
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr10_+_75910908
|
0.895
|
NM_001202450
NM_006721
|
ADK
|
adenosine kinase
|
|
chr3_-_145878775
|
0.890
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr12_+_7023490
|
0.888
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr7_-_72742084
|
0.886
|
NM_178125
|
TRIM50
|
tripartite motif containing 50
|
|
chr18_+_56338818
|
0.885
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr3_+_5229357
|
0.881
|
NM_014674
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr11_-_107729523
|
0.876
|
|
SLC35F2
|
solute carrier family 35, member F2
|
|
chr18_+_56338678
|
0.872
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr10_+_112257624
|
0.872
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr12_-_76424474
|
0.860
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr9_-_94185959
|
0.860
|
|
NFIL3
|
nuclear factor, interleukin 3 regulated
|
|
chr3_+_5229430
|
0.856
|
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1
|
|
chr1_-_20812271
|
0.855
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr19_-_4400470
|
0.854
|
NM_001199943
NM_003025
|
SH3GL1
|
SH3-domain GRB2-like 1
|
|
chr6_+_151646634
|
0.852
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr14_+_55034633
|
0.852
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr9_-_94186140
|
0.850
|
NM_005384
|
NFIL3
|
nuclear factor, interleukin 3 regulated
|
|
chr4_+_186317964
|
0.848
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr5_+_68462836
|
0.848
|
NM_031966
|
CCNB1
|
cyclin B1
|
|
chr15_-_42264687
|
0.848
|
|
EHD4
|
EH-domain containing 4
|
|
chr8_-_30515665
|
0.845
|
NM_002095
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa
|
|
chr17_-_74449220
|
0.840
|
|
UBE2O
|
ubiquitin-conjugating enzyme E2O
|
|
chr8_+_26435339
|
0.837
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr15_+_67358161
|
0.835
|
|
SMAD3
|
SMAD family member 3
|
|
chr4_+_186317837
|
0.831
|
NM_181726
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr8_-_125740650
|
0.831
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr7_-_27135524
|
0.825
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chrX_+_77359768
|
0.825
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr7_+_43622638
|
0.825
|
NM_004760
|
STK17A
|
serine/threonine kinase 17a
|
|
chr3_+_156392369
|
0.819
|
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr13_+_48877950
|
0.808
|
|
RB1
|
retinoblastoma 1
|
|
chr11_+_94501503
|
0.802
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr1_+_65613771
|
0.790
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr7_-_148581377
|
0.784
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr17_+_25621105
|
0.783
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr7_-_97601654
|
0.778
|
|
|
|
|
chr18_+_56338783
|
0.774
|
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr19_+_1407664
|
0.772
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr3_-_57113292
|
0.767
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr18_+_56338617
|
0.762
|
NM_006785
NM_173844
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr1_-_19578030
|
0.758
|
|
KIAA0090
|
KIAA0090
|
|
chr14_-_103995336
|
0.755
|
|
|
|
|
chr1_+_1550794
|
0.754
|
NM_001170686
NM_001170687
NM_001170688
NM_080875
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr1_+_209848994
|
0.752
|
|
G0S2
|
G0/G1switch 2
|
|
chr14_+_65171149
|
0.751
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr11_-_76381787
|
0.751
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr1_-_8938691
|
0.750
|
|
ENO1
|
enolase 1, (alpha)
|
|
chrX_+_77359712
|
0.748
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr22_+_45098091
|
0.747
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr7_-_87505640
|
0.747
|
|
SLC25A40
|
solute carrier family 25, member 40
|
|
chr15_+_68570402
|
0.744
|
|
FEM1B
|
fem-1 homolog b (C. elegans)
|
|
chrX_+_64887593
|
0.742
|
|
MSN
|
moesin
|
|
chr1_-_43833689
|
0.739
|
NM_022821
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr16_+_88870214
|
0.738
|
|
CDT1
|
chromatin licensing and DNA replication factor 1
|
|
chr11_+_72929392
|
0.735
|
NM_176071
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2
|
|
chr6_-_144329342
|
0.733
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr17_-_41622727
|
0.729
|
|
ETV4
|
ets variant 4
|
|
chrX_-_132092224
|
0.719
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr7_+_26331493
|
0.719
|
NM_001199835
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr19_+_11039419
|
0.718
|
NM_138358
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr9_-_72287097
|
0.717
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chrX_-_152939256
|
0.717
|
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr3_-_52273031
|
0.717
|
|
TWF2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila)
|
|
chr19_+_4910339
|
0.715
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr11_-_71159442
|
0.714
|
NM_001163817
NM_001360
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr19_+_45281125
|
0.714
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr4_+_186317892
|
0.713
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr7_+_55086922
|
0.709
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr2_-_97652300
|
0.709
|
NM_001122646
|
FAM178B
|
family with sequence similarity 178, member B
|
|
chr17_+_40610905
|
0.705
|
|
ATP6V0A1
|
ATPase, H+ transporting, lysosomal V0 subunit a1
|
|
chr15_-_70390229
|
0.702
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr12_-_99038401
|
0.697
|
NM_153687
NM_201612
NM_201613
|
IKBIP
|
IKBKB interacting protein
|
|
chr10_-_121356474
|
0.697
|
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr14_+_64971417
|
0.693
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr3_-_50374749
|
0.693
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr4_+_57302448
|
0.692
|
|
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase
|
|
chr3_-_50374875
|
0.691
|
NM_170713
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr11_+_126226095
|
0.690
|
NM_006278
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4
|
|
chr1_+_230202966
|
0.689
|
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2)
|
|
chr8_+_26240522
|
0.689
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr1_-_8938727
|
0.687
|
|
ENO1
|
enolase 1, (alpha)
|
|
chr20_+_42295708
|
0.686
|
NM_002466
|
MYBL2
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 2
|
|
chr7_-_87505453
|
0.686
|
|
SLC25A40
|
solute carrier family 25, member 40
|
|
chr11_-_64014140
|
0.686
|
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chr18_+_12308242
|
0.685
|
NM_032525
|
TUBB6
|
tubulin, beta 6 class V
|
|
chr3_-_179169330
|
0.677
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr10_+_3109708
|
0.676
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr7_-_148581359
|
0.676
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr3_+_122785855
|
0.669
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr19_-_14201230
|
0.669
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr15_-_63673918
|
0.663
|
|
CA12
|
carbonic anhydrase XII
|
|
chr4_-_104119475
|
0.660
|
NM_001813
|
CENPE
|
centromere protein E, 312kDa
|
|
chr14_+_24769116
|
0.656
|
|
|
|
|
chr17_-_80056104
|
0.652
|
NM_004104
|
FASN
|
fatty acid synthase
|
|
chr13_+_114239047
|
0.652
|
NM_007111
|
TFDP1
|
transcription factor Dp-1
|
|
chr9_+_5629110
|
0.651
|
NM_001135920
NM_001206557
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr7_-_148581379
|
0.651
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr15_+_63481788
|
0.648
|
|
RAB8B
|
RAB8B, member RAS oncogene family
|