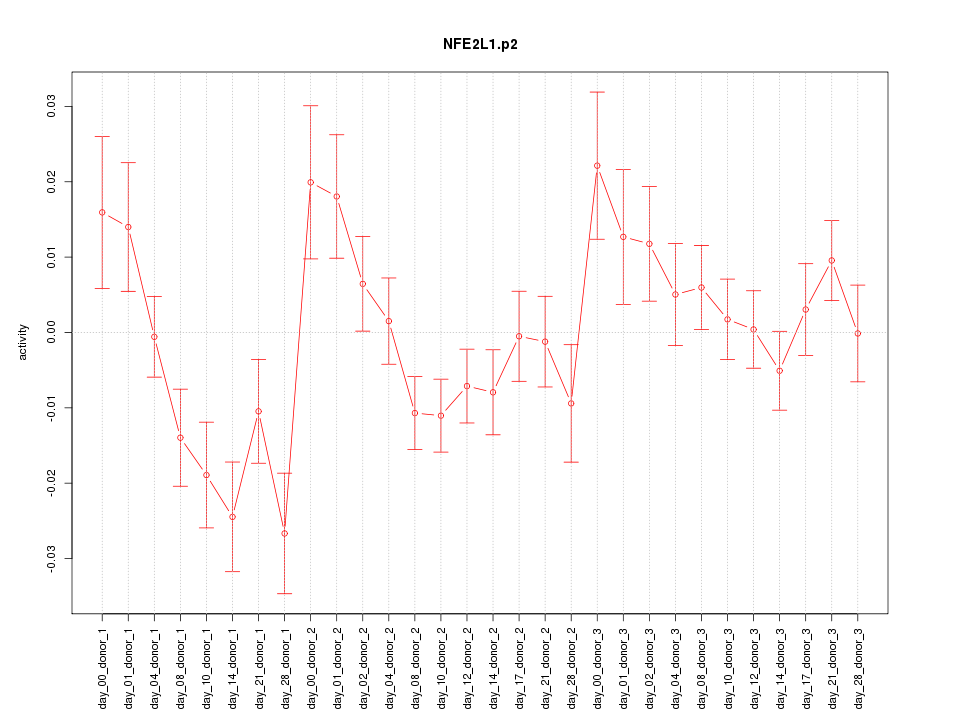
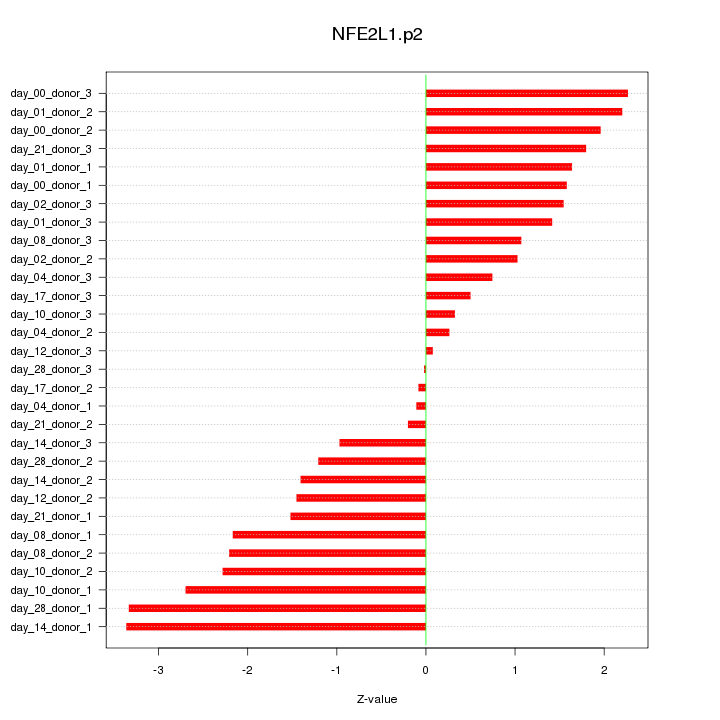
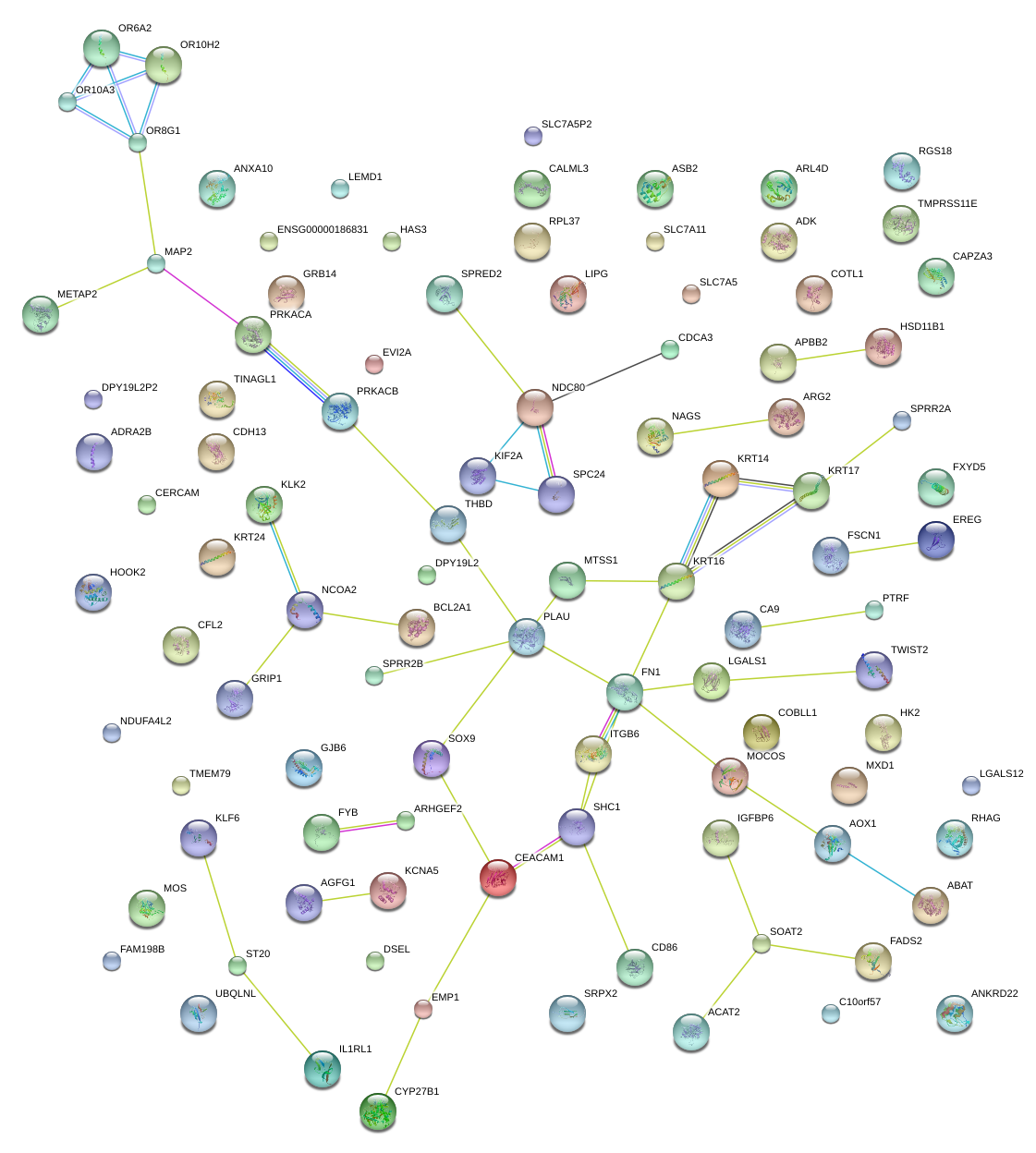
|
chr19_-_36019194
|
11.986
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr2_+_201473942
|
5.516
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr16_+_82660390
|
5.381
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr2_-_161056573
|
5.337
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr2_-_216259197
|
5.040
|
|
FN1
|
fibronectin 1
|
|
chr20_-_23030141
|
4.862
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr17_-_38859976
|
4.757
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr1_-_205391180
|
4.496
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr10_+_5566923
|
4.128
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr13_-_60325073
|
4.093
|
|
|
|
|
chr16_+_82660573
|
4.068
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr14_-_94423766
|
4.035
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr4_-_139163222
|
3.905
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr12_-_123187842
|
3.886
|
NM_177551
|
HCAR2
|
hydroxycarboxylic acid receptor 2
|
|
chr4_+_169013687
|
3.770
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr1_+_32050510
|
3.659
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr12_-_123201319
|
3.570
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr11_+_61595633
|
3.472
|
NM_004265
|
FADS2
|
fatty acid desaturase 2
|
|
chr2_+_210444364
|
3.399
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr7_+_5632435
|
3.306
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr7_+_5632453
|
3.254
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr12_+_13349720
|
3.189
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr17_-_39780762
|
3.131
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr16_+_69139466
|
3.124
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr12_-_57634474
|
3.022
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr17_+_42081913
|
3.005
|
NM_153006
|
NAGS
|
N-acetylglutamate synthase
|
|
chr11_-_5537815
|
2.994
|
NM_145053
|
UBQLNL
|
ubiquilin-like
|
|
chr2_-_165551178
|
2.975
|
|
COBLL1
|
COBL-like 1
|
|
chr15_-_80263460
|
2.937
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr1_-_153044083
|
2.934
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr12_+_13349601
|
2.883
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr2_-_161056762
|
2.795
|
|
ITGB6
|
integrin, beta 6
|
|
chr3_+_121774208
|
2.743
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr2_+_70142303
|
2.693
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr2_+_75061234
|
2.659
|
|
HK2
|
hexokinase 2
|
|
chr10_+_102123420
|
2.641
|
|
|
|
|
chr1_+_156254069
|
2.569
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr1_-_153029987
|
2.520
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr1_-_154943001
|
2.519
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr22_+_38071612
|
2.494
|
NM_002305
|
LGALS1
|
lectin, galactoside-binding, soluble, 1
|
|
chr1_+_84630575
|
2.490
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_+_239756672
|
2.469
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr17_-_39768939
|
2.452
|
NM_005557
|
KRT16
|
keratin 16
|
|
chrX_+_99899162
|
2.433
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr1_+_192127591
|
2.430
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr1_+_84609941
|
2.384
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr4_-_40936640
|
2.339
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr2_-_65593866
|
2.323
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr17_+_70117168
|
2.292
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr16_+_8806825
|
2.280
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr10_+_75936546
|
2.266
|
|
ADK
|
adenosine kinase
|
|
chr13_-_20805371
|
2.264
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr4_-_159092509
|
2.259
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr17_+_70117154
|
2.251
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr18_+_2571509
|
2.196
|
NM_006101
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chrX_+_99899388
|
2.184
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr19_+_35645614
|
2.170
|
NM_014164
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr12_-_67072881
|
2.167
|
NM_001178074
NM_021150
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr1_+_209878135
|
2.162
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr19_-_11266451
|
2.157
|
NM_182513
|
SPC24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr18_-_65182507
|
2.148
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr12_-_6961044
|
2.142
|
|
CDCA3
|
cell division cycle associated 3
|
|
chr19_+_35645640
|
2.137
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr9_+_131191206
|
2.130
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr11_+_61595772
|
2.130
|
|
FADS2
|
fatty acid desaturase 2
|
|
chr18_+_2571587
|
2.115
|
|
NDC80
|
NDC80 kinetochore complex component homolog (S. cerevisiae)
|
|
chr2_-_96781887
|
2.102
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr17_+_70117177
|
2.096
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr11_-_7961066
|
2.095
|
NM_001003745
|
OR10A3
|
olfactory receptor, family 10, subfamily A, member 3
|
|
chr11_-_6817013
|
2.084
|
NM_003696
|
OR6A2
|
olfactory receptor, family 6, subfamily A, member 2
|
|
chr10_-_90611675
|
2.078
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr18_+_47088400
|
2.072
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr12_+_53491417
|
2.063
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr19_+_15838833
|
2.047
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr14_-_35182944
|
2.037
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr10_+_75673346
|
2.028
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr9_+_35673852
|
2.018
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr12_+_53491448
|
1.981
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr17_-_29648631
|
1.974
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr4_+_75230856
|
1.933
|
NM_001432
|
EREG
|
epiregulin
|
|
chr1_-_155939051
|
1.928
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr2_+_210444154
|
1.921
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr2_-_165477818
|
1.877
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr17_-_39743085
|
1.875
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr8_-_57026540
|
1.874
|
NM_005372
|
MOS
|
v-mos Moloney murine sarcoma viral oncogene homolog
|
|
chr2_+_102953716
|
1.849
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr12_-_58160823
|
1.840
|
NM_000785
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr19_-_43031343
|
1.837
|
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr11_+_124120422
|
1.826
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chr10_-_3827205
|
1.820
|
|
KLF6
|
Kruppel-like factor 6
|
|
chr8_-_125580745
|
1.817
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr4_+_69313166
|
1.798
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr16_-_87903028
|
1.793
|
NM_003486
|
SLC7A5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5
|
|
chr16_-_84651639
|
1.781
|
NM_021149
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr12_+_18891040
|
1.778
|
NM_033328
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr6_-_49604524
|
1.766
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr2_+_228336809
|
1.755
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr6_+_160183510
|
1.743
|
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr2_-_165477986
|
1.738
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr7_-_102920665
|
1.729
|
|
DPY19L2P2
|
dpy-19-like 2 pseudogene 2 (C. elegans)
|
|
chr14_+_68086633
|
1.722
|
|
ARG2
|
arginase, type II
|
|
chr11_-_67450536
|
1.707
|
|
RPL37
|
ribosomal protein L37
|
|
chr5_-_39203061
|
1.695
|
|
FYB
|
FYN binding protein
|
|
chr17_-_40575117
|
1.684
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr10_+_82173574
|
1.676
|
NM_001243778
NM_001243782
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr10_+_75936524
|
1.664
|
|
ADK
|
adenosine kinase
|
|
chr12_-_10324675
|
1.664
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr5_-_150948504
|
1.655
|
NM_001447
|
FAT2
|
FAT tumor suppressor homolog 2 (Drosophila)
|
|
chr20_+_20037289
|
1.654
|
NM_001167816
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr12_-_124018066
|
1.654
|
NM_178314
|
RILPL1
|
Rab interacting lysosomal protein-like 1
|
|
chr12_-_89746244
|
1.641
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr11_-_46717657
|
1.641
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr11_-_125366048
|
1.630
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr5_-_112770537
|
1.625
|
NM_032028
|
TSSK1B
|
testis-specific serine kinase 1B
|
|
chr2_-_2334887
|
1.620
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr19_-_51537996
|
1.592
|
NM_019598
NM_145894
NM_145895
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr12_-_89746238
|
1.586
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr12_+_107712151
|
1.575
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr12_-_95611023
|
1.574
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr7_+_134464161
|
1.570
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr3_-_45267068
|
1.569
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr4_-_164534656
|
1.568
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr22_+_22988781
|
1.559
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr16_+_89696775
|
1.549
|
|
DPEP1
|
dipeptidase 1 (renal)
|
|
chr17_+_74381186
|
1.518
|
NM_001142602
|
SPHK1
|
sphingosine kinase 1
|
|
chr8_-_10587942
|
1.517
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr20_+_54823787
|
1.511
|
NM_019888
|
MC3R
|
melanocortin 3 receptor
|
|
chr2_+_113735595
|
1.508
|
NM_019618
|
IL36G
|
interleukin 36, gamma
|
|
chr17_+_39969205
|
1.507
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr4_+_99182520
|
1.506
|
NM_001100426
NM_001100427
NM_001100428
NM_001100429
NM_001100430
NM_021159
|
RAP1GDS1
|
RAP1, GTP-GDP dissociation stimulator 1
|
|
chr17_+_39969200
|
1.500
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr17_+_74381419
|
1.491
|
|
SPHK1
|
sphingosine kinase 1
|
|
chr19_-_11688508
|
1.479
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr18_+_29027731
|
1.475
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr18_-_31628557
|
1.466
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr22_-_37640187
|
1.463
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr4_+_3250766
|
1.461
|
NM_001042690
|
C4orf44
|
chromosome 4 open reading frame 44
|
|
chr11_-_124310980
|
1.457
|
NM_012378
|
OR8B8
|
olfactory receptor, family 8, subfamily B, member 8
|
|
chr4_-_103998169
|
1.451
|
NM_178833
|
SLC9B2
|
solute carrier family 9, subfamily B (cation proton antiporter 2), member 2
|
|
chr12_-_52779362
|
1.451
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr11_-_88781039
|
1.448
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr19_-_11688484
|
1.445
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_-_11688441
|
1.441
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr7_+_134464417
|
1.436
|
|
CALD1
|
caldesmon 1
|
|
chr7_-_41742666
|
1.436
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr4_-_159956332
|
1.431
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr17_+_1959512
|
1.431
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr4_-_159094163
|
1.427
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr15_+_45722762
|
1.417
|
NM_032413
NM_197955
|
C15orf48
|
chromosome 15 open reading frame 48
|
|
chr2_-_31352489
|
1.411
|
NM_001253827
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr2_-_65659115
|
1.407
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr18_+_56530677
|
1.402
|
|
ZNF532
|
zinc finger protein 532
|
|
chr16_-_15833937
|
1.392
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr4_-_68749698
|
1.391
|
NM_004262
|
TMPRSS11D
|
transmembrane protease, serine 11D
|
|
chr17_+_39968961
|
1.388
|
NM_021939
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr14_-_65289812
|
1.383
|
NM_000347
NM_001024858
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr22_-_50523780
|
1.380
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr16_-_72991648
|
1.368
|
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_-_145018904
|
1.366
|
NM_201381
|
PLEC
|
plectin
|
|
chr14_-_94442894
|
1.365
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr1_+_74701070
|
1.362
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr20_+_62795826
|
1.361
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr17_-_39222126
|
1.358
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr5_+_34684611
|
1.355
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr9_-_99180661
|
1.346
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr6_+_160327972
|
1.346
|
NM_002377
|
MAS1
|
MAS1 oncogene
|
|
chr12_-_52585686
|
1.340
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr20_-_33880203
|
1.340
|
NM_178468
|
FAM83C
|
family with sequence similarity 83, member C
|
|
chr19_+_15052298
|
1.337
|
NM_012377
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2
|
|
chr12_-_89745997
|
1.333
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr17_-_7493389
|
1.331
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr9_-_72287097
|
1.323
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr6_-_53213699
|
1.322
|
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr12_-_46633551
|
1.321
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr1_+_152956563
|
1.316
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr17_-_48264039
|
1.316
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr6_-_27880173
|
1.315
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr6_-_53213747
|
1.314
|
NM_001242828
NM_001242830
NM_001242831
NM_021814
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr22_+_42229105
|
1.314
|
NM_004599
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr19_-_42931500
|
1.309
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr1_-_28503454
|
1.296
|
NM_000952
NM_001164723
|
PTAFR
|
platelet-activating factor receptor
|
|
chrX_+_135251795
|
1.296
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_+_57310113
|
1.293
|
NM_001105565
|
SMTNL1
|
smoothelin-like 1
|
|
chr3_-_48040260
|
1.291
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr2_-_193059249
|
1.285
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr15_+_74165967
|
1.284
|
NM_153356
|
TBC1D21
|
TBC1 domain family, member 21
|
|
chr2_+_172950207
|
1.284
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr3_-_50374795
|
1.273
|
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1
|
|
chr14_-_94443065
|
1.273
|
NM_001202429
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr10_+_75936443
|
1.272
|
|
ADK
|
adenosine kinase
|
|
chr19_+_11200124
|
1.272
|
|
LDLR
|
low density lipoprotein receptor
|
|
chr10_-_23003443
|
1.262
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr11_+_66188474
|
1.260
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr22_-_50523737
|
1.253
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr6_+_151646634
|
1.249
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr9_+_115631814
|
1.247
|
|
SNX30
|
sorting nexin family member 30
|
|
chr5_+_137514753
|
1.246
|
|
KIF20A
|
kinesin family member 20A
|
|
chr2_-_47134945
|
1.242
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr11_-_87908551
|
1.236
|
NM_022337
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr14_+_68086655
|
1.234
|
|
ARG2
|
arginase, type II
|