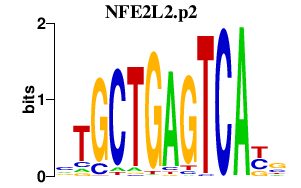
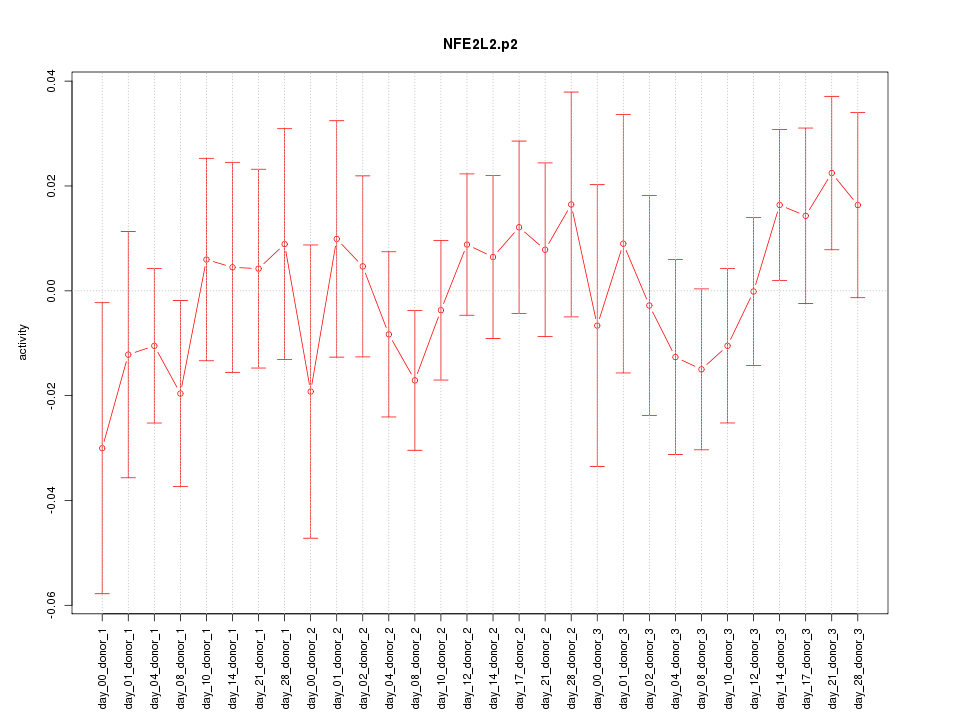
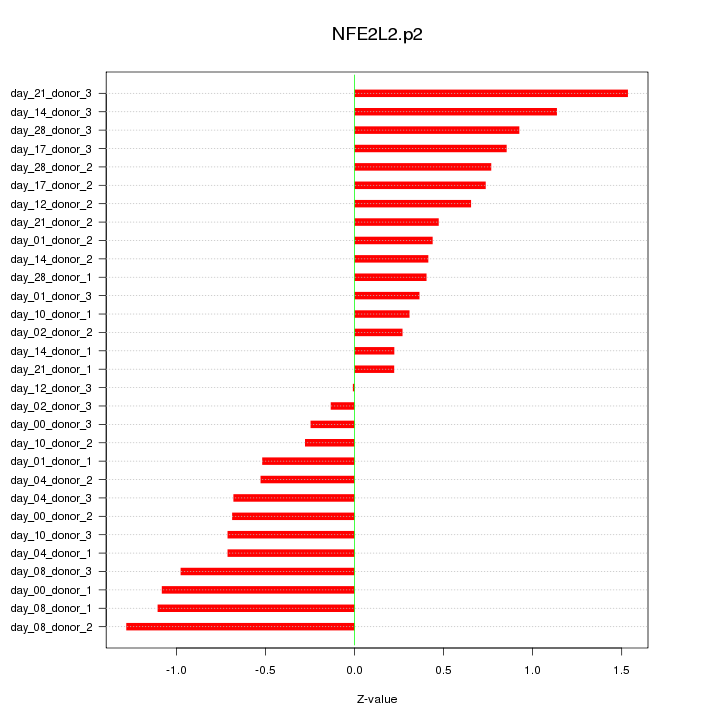
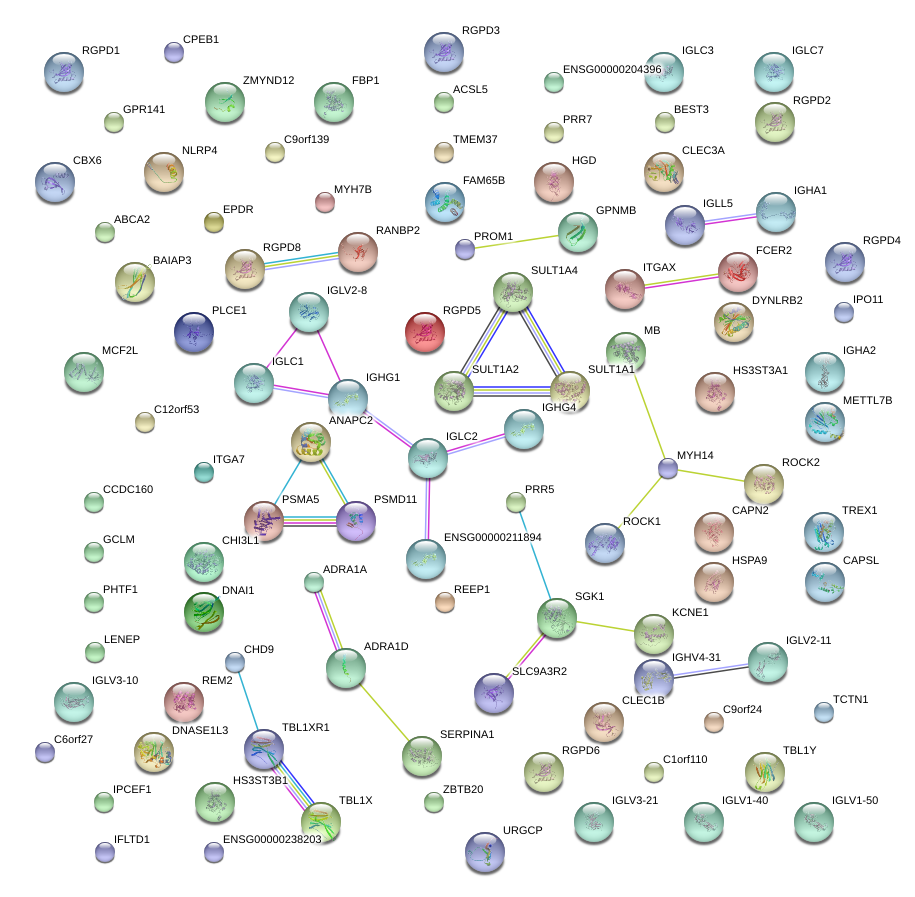
|
chr9_-_34397786
|
3.327
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr7_+_23286384
|
1.886
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr12_+_56075329
|
1.799
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr7_+_37960418
|
1.772
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr12_+_56075418
|
1.688
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr7_+_23286277
|
1.677
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_+_37960920
|
1.645
|
NM_001242948
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr4_-_16077740
|
1.612
|
NM_001145849
NM_001145850
NM_001145851
NM_001145852
NM_006017
|
PROM1
|
prominin 1
|
|
chr19_+_50706868
|
1.509
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr7_+_23286386
|
1.488
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_-_162838550
|
1.429
|
NM_178550
|
C1orf110
|
chromosome 1 open reading frame 110
|
|
chr7_+_23286390
|
1.241
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr14_-_21572374
|
1.224
|
|
|
|
|
chr16_+_80574853
|
1.145
|
NM_130897
|
DYNLRB2
|
dynein, light chain, roadblock-type 2
|
|
chr5_-_35938736
|
1.132
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr9_+_140135738
|
1.130
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr10_+_114136014
|
1.111
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr16_+_2083264
|
1.107
|
NM_001252073
NM_001252075
NM_001252076
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr19_+_56347943
|
1.082
|
NM_134444
|
NLRP4
|
NLR family, pyrin domain containing 4
|
|
chr9_+_139921915
|
1.045
|
NM_207511
|
C9orf139
|
chromosome 9 open reading frame 139
|
|
chr16_-_28620648
|
1.025
|
NM_177530
NM_177534
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr9_+_140135698
|
0.978
|
NM_006088
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr14_-_106926393
|
0.915
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_+_53133063
|
0.906
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr2_+_120189342
|
0.883
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr16_-_28607694
|
0.846
|
NM_177528
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr1_-_94374986
|
0.775
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr14_-_106641585
|
0.768
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr3_-_120401401
|
0.765
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr9_+_34458810
|
0.757
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr1_+_223889253
|
0.753
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr8_-_26724748
|
0.748
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chrX_+_133371076
|
0.746
|
NM_001101357
|
CCDC160
|
coiled-coil domain containing 160
|
|
chr3_-_120400957
|
0.725
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr5_+_176875325
|
0.716
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr1_-_203155813
|
0.704
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr13_+_113698907
|
0.690
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_+_45072936
|
0.686
|
|
PRR5
|
proline rich 5 (renal)
|
|
chr14_+_23352430
|
0.682
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr10_+_114135937
|
0.682
|
NM_016234
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr14_-_94856926
|
0.680
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr12_-_10151689
|
0.680
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr1_+_154966061
|
0.671
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr8_-_11204998
|
0.670
|
|
LOC100129129
|
uncharacterized LOC100129129
|
|
chr9_-_139922705
|
0.647
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr9_-_139922595
|
0.647
|
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr1_-_114302093
|
0.646
|
|
PHTF1
|
putative homeodomain transcription factor 1
|
|
chr16_+_78056411
|
0.632
|
NM_005752
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr2_-_111334677
|
0.630
|
NM_005054
|
RGPD5
|
RANBP2-like and GRIP domain containing 5
|
|
chr1_-_109969058
|
0.630
|
NM_001199772
NM_002790
NM_001199773
NM_001199774
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr16_-_28621279
|
0.629
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr7_+_23286420
|
0.626
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr6_-_134496833
|
0.617
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_-_106774311
|
0.615
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr9_-_97402530
|
0.605
|
NM_001127628
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr16_+_31366529
|
0.600
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr22_+_22735202
|
0.586
|
|
|
|
|
chr10_+_95848811
|
0.583
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr10_+_114135965
|
0.579
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr3_-_114343791
|
0.575
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr15_-_83240499
|
0.551
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr16_+_1386197
|
0.549
|
NM_001199096
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr12_-_25801487
|
0.535
|
NM_001145727
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr16_+_31366519
|
0.526
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr19_-_7764283
|
0.526
|
NM_001207019
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr2_+_110551965
|
0.524
|
NM_005054
|
RGPD5
|
RANBP2-like and GRIP domain containing 5
|
|
chr22_+_45072687
|
0.524
|
NM_001017530
NM_001198721
NM_015366
|
PRR5
|
proline rich 5 (renal)
|
|
chr17_+_14204326
|
0.513
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr12_-_56101667
|
0.499
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr5_+_61714710
|
0.490
|
NM_001134779
|
IPO11
|
importin 11
|
|
chrX_+_9431314
|
0.489
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr12_-_6809965
|
0.486
|
NM_001244014
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chr6_-_31745048
|
0.483
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr1_-_94374949
|
0.475
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr3_+_48507621
|
0.459
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr9_-_140083034
|
0.458
|
NM_013366
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr22_-_36013303
|
0.457
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr22_+_23040394
|
0.456
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr3_-_58200397
|
0.453
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr21_-_35828038
|
0.446
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr2_-_86565205
|
0.444
|
NM_001164730
|
REEP1
|
receptor accessory protein 1
|
|
chr12_-_56101465
|
0.441
|
|
ITGA7
|
integrin, alpha 7
|
|
chr2_-_113191106
|
0.440
|
NM_001164463
NM_005054
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr9_-_97402556
|
0.433
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr1_-_42921676
|
0.423
|
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr2_-_11484699
|
0.416
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr3_+_48507209
|
0.416
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr12_-_70083055
|
0.413
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr22_-_39268214
|
0.410
|
|
CBX6
|
chromobox homolog 6
|
|
chr12_+_111051831
|
0.407
|
NM_001082537
NM_001082538
NM_001173976
NM_024549
NM_001173975
|
TCTN1
|
tectonic family member 1
|
|
chr6_-_24877453
|
0.404
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr6_-_154677868
|
0.404
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr17_+_30771522
|
0.401
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr5_-_137911039
|
0.397
|
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr17_+_30771485
|
0.396
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr3_+_184038445
|
0.395
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr6_-_134497069
|
0.394
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_+_231921625
|
0.393
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr7_+_116312412
|
0.393
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr17_+_30771494
|
0.392
|
NM_002815
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr17_+_30771532
|
0.392
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr12_-_15114495
|
0.386
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chrX_-_53310748
|
0.383
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr22_+_38035447
|
0.383
|
|
|
|
|
chr7_+_23286401
|
0.383
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr2_-_107084800
|
0.380
|
NM_001144013
|
RGPD3
|
RANBP2-like and GRIP domain containing 3
|
|
chr1_+_14075931
|
0.380
|
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr9_-_140082983
|
0.379
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr19_+_13135809
|
0.376
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr2_+_69201704
|
0.376
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr11_+_10477480
|
0.374
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr9_+_140135742
|
0.374
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr22_+_22723972
|
0.372
|
|
|
|
|
chr3_+_184038050
|
0.371
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr22_-_27013959
|
0.371
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr3_-_39234071
|
0.371
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr5_-_137911005
|
0.367
|
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr17_+_30771520
|
0.366
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr5_+_179247911
|
0.361
|
|
SQSTM1
|
sequestosome 1
|
|
chr19_+_13135368
|
0.360
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr11_+_10476665
|
0.358
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr14_-_106967528
|
0.349
|
|
IGHA1
IGHA2
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr17_+_79651052
|
0.345
|
|
|
|
|
chr8_-_82395401
|
0.345
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr16_-_30124829
|
0.341
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr1_+_14075917
|
0.339
|
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr1_-_42921923
|
0.336
|
NM_001146192
NM_032257
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr11_+_35684418
|
0.334
|
|
TRIM44
|
tripartite motif containing 44
|
|
chr11_+_35684342
|
0.330
|
NM_017583
|
TRIM44
|
tripartite motif containing 44
|
|
chr9_-_116840604
|
0.330
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr19_-_4559713
|
0.330
|
NM_032108
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr16_+_22308722
|
0.328
|
NM_018119
|
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD)
|
|
chr3_-_170587947
|
0.327
|
NM_001099645
|
RPL22L1
|
ribosomal protein L22-like 1
|
|
chr22_+_22735134
|
0.323
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr1_-_94375028
|
0.322
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr18_-_267987
|
0.318
|
NM_005131
|
THOC1
|
THO complex 1
|
|
chrX_-_110513750
|
0.318
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr19_+_41430123
|
0.315
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr22_+_22735207
|
0.314
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr3_-_150920946
|
0.310
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr1_+_27719151
|
0.309
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr1_-_177939347
|
0.306
|
|
SEC16B
|
SEC16 homolog B (S. cerevisiae)
|
|
chr10_+_695887
|
0.305
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr2_+_18059923
|
0.304
|
NM_002252
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3
|
|
chr10_-_13544944
|
0.298
|
NM_001100912
NM_152751
|
BEND7
|
BEN domain containing 7
|
|
chr22_+_22569155
|
0.295
|
|
|
|
|
chr7_-_102985068
|
0.292
|
NM_001129887
NM_014377
|
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2
|
|
chr1_-_93257950
|
0.292
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr14_-_24777064
|
0.290
|
|
|
|
|
chr17_-_72889704
|
0.289
|
NM_178128
|
FADS6
|
fatty acid desaturase domain family, member 6
|
|
chr11_-_62474810
|
0.287
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr11_-_65150107
|
0.286
|
NM_001077241
NM_182556
|
SLC25A45
|
solute carrier family 25, member 45
|
|
chr1_+_22351984
|
0.286
|
|
LINC00339
|
long intergenic non-protein coding RNA 339
|
|
chr18_+_61637262
|
0.284
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr5_+_179247903
|
0.284
|
|
SQSTM1
|
sequestosome 1
|
|
chr3_+_183903970
|
0.278
|
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chrX_-_153210057
|
0.278
|
NM_002910
|
RENBP
|
renin binding protein
|
|
chr10_+_85899184
|
0.277
|
NM_014394
|
GHITM
|
growth hormone inducible transmembrane protein
|
|
chr18_-_40695483
|
0.275
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chr1_-_114301753
|
0.274
|
NM_006608
|
PHTF1
|
putative homeodomain transcription factor 1
|
|
chr2_-_219134795
|
0.273
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr16_+_57662301
|
0.271
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr3_+_183903862
|
0.270
|
NM_018358
|
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3
|
|
chr2_-_238499317
|
0.267
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr17_-_39334439
|
0.266
|
NM_033062
|
KRTAP4-2
|
keratin associated protein 4-2
|
|
chr22_-_27014037
|
0.264
|
|
CRYBB1
|
crystallin, beta B1
|
|
chr17_+_73598139
|
0.264
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr19_+_3366589
|
0.263
|
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr18_-_58040000
|
0.263
|
NM_005912
|
MC4R
|
melanocortin 4 receptor
|
|
chr5_-_149535420
|
0.263
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr10_-_123356158
|
0.263
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr19_-_49658645
|
0.262
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr9_-_116840551
|
0.260
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr1_+_114493766
|
0.260
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr22_-_39268230
|
0.258
|
NM_014292
|
CBX6
|
chromobox homolog 6
|
|
chr2_-_219134865
|
0.258
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr12_-_71182699
|
0.257
|
NM_001207015
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr2_-_28113954
|
0.257
|
|
LOC100302650
|
uncharacterized LOC100302650
|
|
chr11_-_62474666
|
0.256
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr8_-_143823823
|
0.255
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr16_-_66959418
|
0.255
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr16_+_30419363
|
0.254
|
|
ZNF771
|
zinc finger protein 771
|
|
chr6_+_44195008
|
0.254
|
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr16_+_71560022
|
0.252
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chr9_+_132251403
|
0.251
|
|
LOC100506190
|
uncharacterized LOC100506190
|
|
chr22_+_22764094
|
0.251
|
|
|
|
|
chr2_-_219134846
|
0.250
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr14_-_23284565
|
0.249
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr17_+_2497233
|
0.248
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr7_+_150434435
|
0.247
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr2_+_109335906
|
0.243
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr19_+_13135745
|
0.242
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr16_-_27899477
|
0.241
|
NM_144675
|
GSG1L
|
GSG1-like
|
|
chrX_-_64754594
|
0.238
|
|
LAS1L
|
LAS1-like (S. cerevisiae)
|
|
chr14_-_23284617
|
0.238
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr7_-_1498961
|
0.237
|
|
MICALL2
|
MICAL-like 2
|
|
chr9_+_130922471
|
0.237
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr3_+_132373289
|
0.236
|
NM_198329
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr17_+_48610077
|
0.236
|
|
EPN3
|
epsin 3
|
|
chr6_-_56258891
|
0.233
|
|
COL21A1
|
collagen, type XXI, alpha 1
|