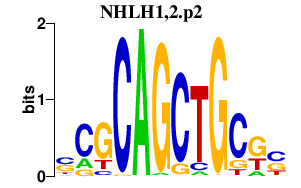
|
chr3_-_122746544
|
2.119
|
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B
|
|
chr2_-_7005764
|
1.845
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr13_-_26625164
|
1.765
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr4_+_75858205
|
1.560
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr17_-_39280377
|
1.467
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr16_+_67465019
|
1.435
|
NM_000196
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2
|
|
chr11_+_113930430
|
1.050
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr8_-_17271015
|
0.919
|
NM_004686
|
MTMR7
|
myotubularin related protein 7
|
|
chr6_-_90121966
|
0.862
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr19_+_18111923
|
0.858
|
NM_001025604
|
ARRDC2
|
arrestin domain containing 2
|
|
chr17_-_39316982
|
0.835
|
NM_032524
|
KRTAP4-4
|
keratin associated protein 4-4
|
|
chr6_-_90121656
|
0.832
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr10_+_94833646
|
0.802
|
NM_000783
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr12_-_63328857
|
0.795
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr12_+_102091399
|
0.793
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr17_+_39261640
|
0.776
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr12_-_63328663
|
0.767
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr21_+_42688660
|
0.749
|
NM_058186
NM_206964
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr6_-_46138584
|
0.736
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr10_-_13043570
|
0.734
|
NM_031455
|
CCDC3
|
coiled-coil domain containing 3
|
|
chr12_-_29936685
|
0.708
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr17_-_39334439
|
0.688
|
NM_033062
|
KRTAP4-2
|
keratin associated protein 4-2
|
|
chr5_+_156693051
|
0.687
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr19_-_14316980
|
0.680
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr6_-_90121911
|
0.677
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr11_+_113930296
|
0.663
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr6_+_126112057
|
0.659
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr1_-_22469414
|
0.651
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr5_+_76114832
|
0.648
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr7_-_139762339
|
0.635
|
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr5_+_76114881
|
0.623
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr14_-_65438794
|
0.620
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chrX_+_68725077
|
0.612
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr2_+_10861740
|
0.602
|
NM_001039362
NM_144583
|
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2
|
|
chr7_+_17338182
|
0.602
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr11_-_46867788
|
0.591
|
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr6_-_154831652
|
0.588
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr17_+_39382878
|
0.584
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chrX_+_16964730
|
0.577
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr16_-_55866972
|
0.575
|
|
CES1
|
carboxylesterase 1
|
|
chr7_-_131241321
|
0.565
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr6_-_154831788
|
0.564
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr5_-_94620277
|
0.558
|
NM_024717
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr14_+_73957608
|
0.550
|
NM_024644
|
C14orf169
|
chromosome 14 open reading frame 169
|
|
chr17_-_27278324
|
0.546
|
|
PHF12
|
PHD finger protein 12
|
|
chr2_-_9563584
|
0.545
|
|
ITGB1BP1
|
integrin beta 1 binding protein 1
|
|
chr20_+_44637539
|
0.542
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr3_-_195306274
|
0.540
|
|
APOD
|
apolipoprotein D
|
|
chr20_+_58179601
|
0.536
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr20_+_37434294
|
0.534
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr17_+_72732958
|
0.516
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr3_-_45267068
|
0.515
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr14_+_73957657
|
0.513
|
|
C14orf169
|
chromosome 14 open reading frame 169
|
|
chr14_+_73957685
|
0.510
|
|
C14orf169
|
chromosome 14 open reading frame 169
|
|
chr17_-_4689508
|
0.510
|
NM_001144939
NM_001144940
NM_001144941
NM_182566
|
VMO1
|
vitelline membrane outer layer 1 homolog (chicken)
|
|
chr1_+_233749917
|
0.509
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr10_-_42863435
|
0.504
|
|
LOC441666
|
zinc finger protein 91 pseudogene
|
|
chr14_+_73957670
|
0.485
|
|
C14orf169
|
chromosome 14 open reading frame 169
|
|
chr16_-_55867016
|
0.483
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr8_-_17270800
|
0.481
|
|
MTMR7
|
myotubularin related protein 7
|
|
chr11_+_46299554
|
0.471
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr16_+_330605
|
0.469
|
NM_001176
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma
|
|
chr6_+_147524866
|
0.464
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr6_-_154831704
|
0.460
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr12_+_90102639
|
0.458
|
|
LOC338758
|
uncharacterized LOC338758
|
|
chr11_-_66335958
|
0.445
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr1_-_38273852
|
0.441
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr21_+_34442550
|
0.434
|
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr1_-_38273823
|
0.425
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr21_+_27011588
|
0.423
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr15_+_41062158
|
0.423
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr2_-_233352530
|
0.422
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr5_+_67511578
|
0.418
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr20_-_3154169
|
0.417
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr8_+_17014006
|
0.416
|
|
ZDHHC2
|
zinc finger, DHHC-type containing 2
|
|
chr22_+_29279573
|
0.412
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr15_-_27018216
|
0.410
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr6_-_46703042
|
0.408
|
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr9_+_37650996
|
0.404
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr1_+_87797371
|
0.404
|
|
LMO4
|
LIM domain only 4
|
|
chr11_+_94277566
|
0.400
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr1_+_87794548
|
0.399
|
|
LMO4
|
LIM domain only 4
|
|
chr1_+_218519576
|
0.392
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr10_-_135150474
|
0.391
|
NM_015722
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr5_+_43603312
|
0.386
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chrX_-_63425484
|
0.385
|
NM_152424
|
FAM123B
|
family with sequence similarity 123B
|
|
chr5_+_43603216
|
0.382
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr13_-_36705432
|
0.381
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr11_-_207324
|
0.381
|
|
BET1L
|
blocked early in transport 1 homolog (S. cerevisiae)-like
|
|
chr6_-_167040700
|
0.380
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr7_-_137531585
|
0.380
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr19_+_3094528
|
0.379
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr4_-_149363498
|
0.377
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr1_+_87794150
|
0.377
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr3_-_42305438
|
0.372
|
NM_001174138
|
CCK
|
cholecystokinin
|
|
chr16_-_69373406
|
0.371
|
|
COG8
|
component of oligomeric golgi complex 8
|
|
chr4_+_41362750
|
0.362
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr9_-_75568232
|
0.361
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr10_-_135150368
|
0.360
|
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr9_-_112970412
|
0.358
|
NM_001012993
|
C9orf152
|
chromosome 9 open reading frame 152
|
|
chr1_-_38273846
|
0.354
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr20_+_42086533
|
0.354
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr15_-_27018917
|
0.353
|
NM_021912
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr10_+_123923104
|
0.352
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_+_27062508
|
0.351
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr13_+_35516390
|
0.351
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr5_+_141016987
|
0.350
|
|
RELL2
|
RELT-like 2
|
|
chr10_+_123922940
|
0.346
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr3_-_120068077
|
0.346
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr16_+_30406711
|
0.342
|
NM_152652
|
ZNF48
|
zinc finger protein 48
|
|
chr17_-_27278471
|
0.342
|
NM_001033561
NM_020889
|
PHF12
|
PHD finger protein 12
|
|
chr1_+_38273817
|
0.341
|
|
C1orf122
|
chromosome 1 open reading frame 122
|
|
chr12_-_120554558
|
0.340
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr8_-_16859540
|
0.338
|
NM_019851
|
FGF20
|
fibroblast growth factor 20
|
|
chr1_+_233749749
|
0.337
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr17_-_72968750
|
0.331
|
|
|
|
|
chr14_-_89020888
|
0.329
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr16_+_19179534
|
0.327
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr21_+_36041687
|
0.327
|
NM_053277
|
CLIC6
|
chloride intracellular channel 6
|
|
chr5_+_141016567
|
0.325
|
|
RELL2
|
RELT-like 2
|
|
chr10_+_93170086
|
0.324
|
NM_173497
NM_182765
|
HECTD2
|
HECT domain containing 2
|
|
chr6_-_167040659
|
0.323
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr1_+_229761988
|
0.321
|
|
URB2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae)
|
|
chr8_-_98290072
|
0.321
|
|
TSPYL5
|
TSPY-like 5
|
|
chr3_+_69788562
|
0.316
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_+_71320105
|
0.314
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr17_+_3539761
|
0.312
|
NM_001031681
NM_004937
|
CTNS
|
cystinosin, lysosomal cystine transporter
|
|
chr1_-_117753476
|
0.307
|
NM_001253850
NM_024626
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr12_+_56137063
|
0.306
|
NM_005811
|
GDF11
|
growth differentiation factor 11
|
|
chr1_-_47184790
|
0.306
|
|
|
|
|
chr5_+_141016516
|
0.306
|
NM_001130029
NM_173828
|
RELL2
|
RELT-like 2
|
|
chr2_+_97481977
|
0.303
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr1_+_46269283
|
0.302
|
NM_015112
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr8_-_98290171
|
0.300
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr16_-_69373473
|
0.300
|
NM_032382
|
COG8
|
component of oligomeric golgi complex 8
|
|
chr9_+_90340973
|
0.299
|
NM_001912
NM_145918
|
CTSL1
|
cathepsin L1
|
|
chr1_-_6662681
|
0.299
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr15_-_78369825
|
0.297
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr5_+_43603287
|
0.293
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr5_+_67511601
|
0.292
|
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr7_+_12250818
|
0.292
|
NM_001134232
NM_018374
|
TMEM106B
|
transmembrane protein 106B
|
|
chr9_+_132251403
|
0.292
|
|
LOC100506190
|
uncharacterized LOC100506190
|
|
chr17_+_80693398
|
0.291
|
NM_022158
|
FN3K
|
fructosamine 3 kinase
|
|
chr17_+_72733350
|
0.290
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr6_-_35696359
|
0.287
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr7_-_73184535
|
0.285
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr2_+_191745912
|
0.284
|
|
GLS
|
glutaminase
|
|
chr7_+_23636970
|
0.284
|
NM_138771
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr7_-_95225802
|
0.283
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr7_-_120498009
|
0.282
|
|
TSPAN12
|
tetraspanin 12
|
|
chr11_+_62104773
|
0.282
|
NM_001083926
NM_025080
|
ASRGL1
|
asparaginase like 1
|
|
chr16_-_66730466
|
0.281
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr3_+_30648372
|
0.281
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr16_+_58059469
|
0.281
|
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr1_+_87794576
|
0.280
|
|
LMO4
|
LIM domain only 4
|
|
chr13_-_114566951
|
0.280
|
|
GAS6
|
growth arrest-specific 6
|
|
chr19_+_4304590
|
0.279
|
NM_024333
|
FSD1
|
fibronectin type III and SPRY domain containing 1
|
|
chr3_-_48470715
|
0.278
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr19_+_17416476
|
0.276
|
NM_023937
|
MRPL34
|
mitochondrial ribosomal protein L34
|
|
chr2_+_85980600
|
0.276
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr9_-_104357202
|
0.275
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr12_+_93771607
|
0.274
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr19_+_18118921
|
0.274
|
NM_015683
|
ARRDC2
|
arrestin domain containing 2
|
|
chr3_-_195538699
|
0.273
|
NM_004532
NM_018406
NM_138297
|
MUC4
|
mucin 4, cell surface associated
|
|
chr16_+_53133063
|
0.273
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr16_-_49315682
|
0.273
|
NM_004352
|
CBLN1
|
cerebellin 1 precursor
|
|
chr7_-_2883649
|
0.272
|
NM_007353
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12
|
|
chr4_-_1685717
|
0.271
|
NM_001013622
NM_001174070
|
FAM53A
|
family with sequence similarity 53, member A
|
|
chr11_+_62104882
|
0.270
|
|
ASRGL1
|
asparaginase like 1
|
|
chr22_-_18923654
|
0.270
|
|
PRODH
|
proline dehydrogenase (oxidase) 1
|
|
chr16_+_83932729
|
0.268
|
NM_012213
|
MLYCD
|
malonyl-CoA decarboxylase
|
|
chr9_+_74764394
|
0.268
|
|
GDA
|
guanine deaminase
|
|
chr1_+_1167622
|
0.268
|
NM_080605
|
B3GALT6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6
|
|
chr1_+_151584514
|
0.265
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr16_+_22825481
|
0.265
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr12_+_120554692
|
0.264
|
|
|
|
|
chr13_-_29292961
|
0.263
|
NM_001135919
NM_181785
|
SLC46A3
|
solute carrier family 46, member 3
|
|
chr12_-_120554453
|
0.262
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr4_+_7045142
|
0.262
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr16_+_22825806
|
0.261
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr8_-_110657745
|
0.260
|
NM_001099751
NM_001099752
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr17_+_3540004
|
0.260
|
|
CTNS
|
cystinosin, lysosomal cystine transporter
|
|
chr9_+_74764382
|
0.257
|
|
GDA
|
guanine deaminase
|
|
chr6_-_32551993
|
0.257
|
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr5_+_149109850
|
0.256
|
|
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta
|
|
chr1_-_32403975
|
0.255
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr15_-_27018160
|
0.254
|
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr3_-_133614421
|
0.254
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr1_+_25943958
|
0.253
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr14_+_74003817
|
0.253
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr1_-_177133818
|
0.253
|
|
ASTN1
|
astrotactin 1
|
|
chr9_+_90341123
|
0.252
|
|
CTSL1
|
cathepsin L1
|
|
chr1_+_151584665
|
0.251
|
|
SNX27
|
sorting nexin family member 27
|
|
chr17_+_14204326
|
0.251
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr8_+_42995628
|
0.251
|
|
HGSNAT
|
heparan-alpha-glucosaminide N-acetyltransferase
|
|
chr7_+_45197366
|
0.250
|
NM_005856
|
RAMP3
|
receptor (G protein-coupled) activity modifying protein 3
|
|
chr7_+_150026877
|
0.249
|
NM_138434
|
C7orf29
|
chromosome 7 open reading frame 29
|
|
chr1_-_144994943
|
0.248
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_144994908
|
0.248
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr3_-_118753665
|
0.248
|
NM_001015887
|
IGSF11
|
immunoglobulin superfamily, member 11
|