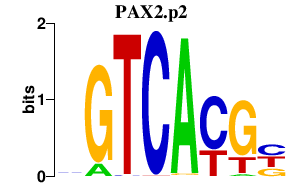
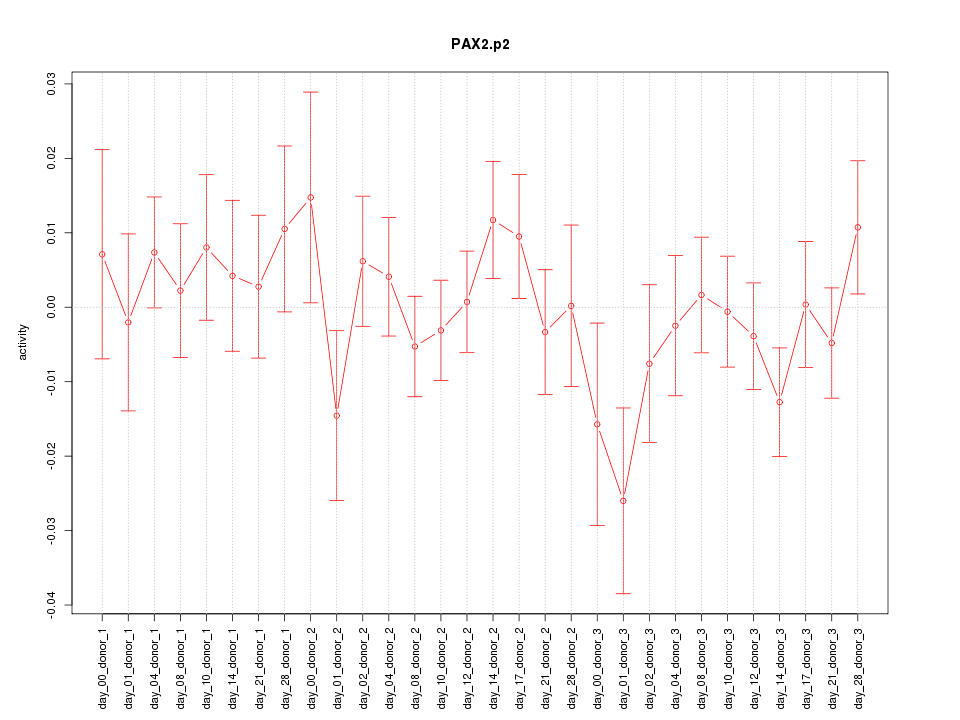
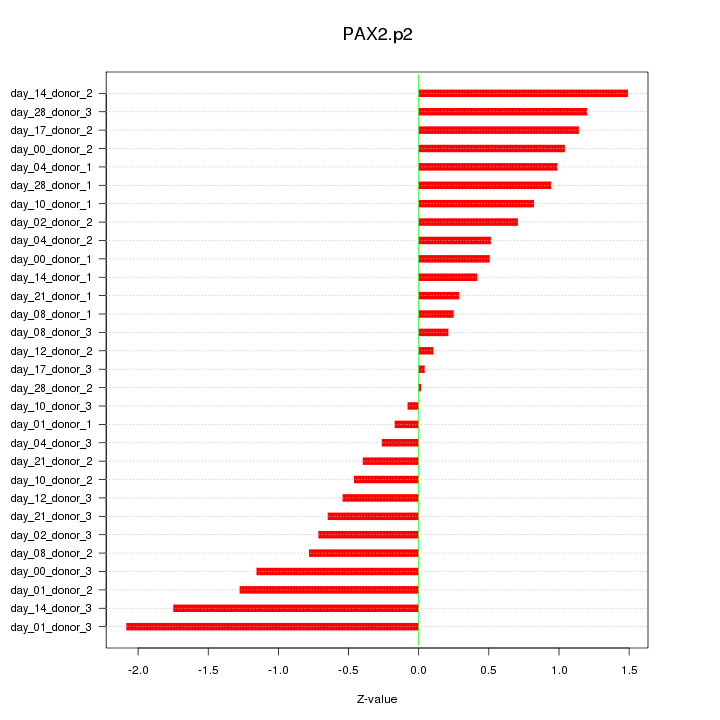
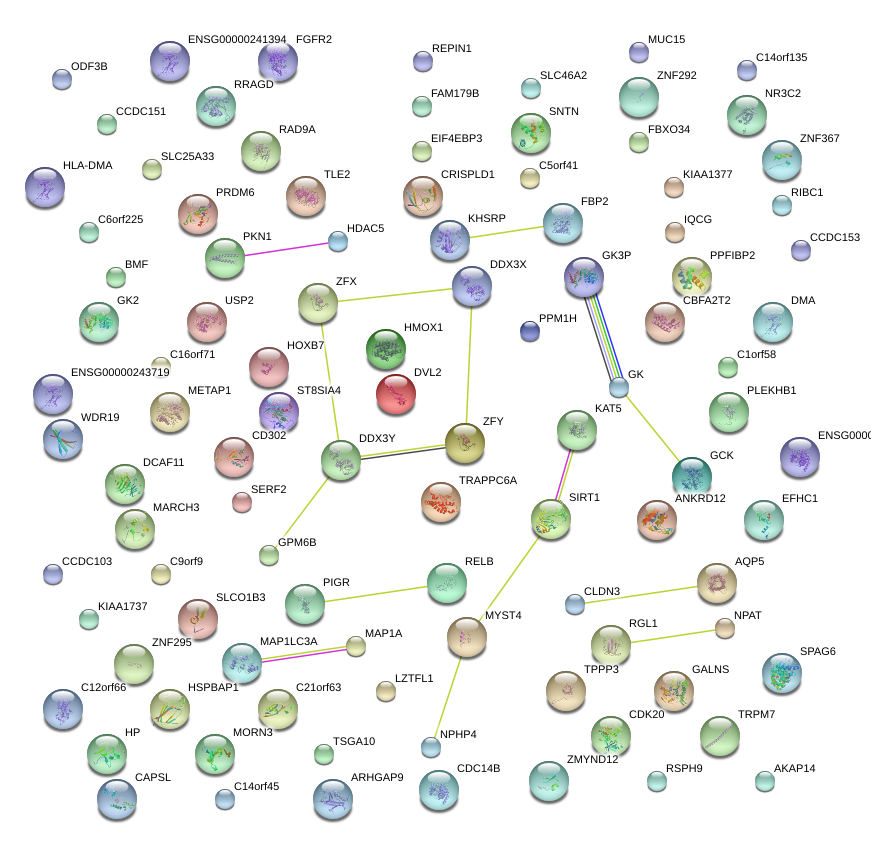
|
chr1_-_42921676
|
2.409
|
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr9_-_115652961
|
1.328
|
NM_033051
|
SLC46A2
|
solute carrier family 46, member 2
|
|
chr11_-_119252267
|
1.255
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr10_+_22634373
|
1.252
|
NM_001253854
NM_001253855
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr11_+_65479685
|
1.209
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr1_-_42921923
|
1.208
|
NM_001146192
NM_032257
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr6_+_112408673
|
1.114
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr16_-_67427335
|
1.094
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr10_+_69644341
|
1.083
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr5_+_172483285
|
1.062
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr11_+_65479472
|
1.056
|
NM_001206833
NM_006388
NM_182709
NM_182710
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr3_+_63638343
|
0.987
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr1_-_207119707
|
0.969
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr3_-_197686837
|
0.945
|
NM_032263
|
IQCG
|
IQ motif containing G
|
|
chr14_+_77564570
|
0.941
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr11_-_26593667
|
0.925
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr16_-_88923233
|
0.898
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr11_+_73358593
|
0.898
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr15_+_43809805
|
0.876
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chrY_+_21729214
|
0.861
|
|
TXLNG2P
|
taxilin gamma 2, pseudogene
|
|
chr11_-_119252364
|
0.858
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_+_67159421
|
0.854
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr5_-_35938736
|
0.843
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chrY_+_15016698
|
0.838
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr2_-_160654668
|
0.831
|
|
CD302
|
CD302 molecule
|
|
chrX_+_119029799
|
0.823
|
NM_001008534
NM_001008535
NM_178813
|
AKAP14
|
A kinase (PRKA) anchor protein 14
|
|
chr22_-_50970542
|
0.808
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr9_-_90589360
|
0.792
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr19_-_45681454
|
0.789
|
NM_024108
|
TRAPPC6A
|
trafficking protein particle complex 6A
|
|
chr14_+_24584009
|
0.755
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr19_-_3047632
|
0.753
|
NM_001144761
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr17_-_42200957
|
0.752
|
|
HDAC5
|
histone deacetylase 5
|
|
chr8_+_75896707
|
0.747
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr19_+_45504686
|
0.746
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chrX_+_30671456
|
0.736
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr16_+_84178864
|
0.732
|
NM_178452
|
DNAAF1
|
dynein, axonemal, assembly factor 1
|
|
chr11_-_108093262
|
0.724
|
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chr1_+_222885895
|
0.723
|
NM_144695
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr1_-_6052481
|
0.712
|
NM_015102
|
NPHP4
|
nephronophthisis 4
|
|
chr1_-_6052410
|
0.707
|
|
NPHP4
|
nephronophthisis 4
|
|
chr12_+_50355278
|
0.702
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr14_+_74486049
|
0.696
|
NM_025057
|
C14orf45
|
chromosome 14 open reading frame 45
|
|
chrX_+_30671602
|
0.693
|
|
GK
|
glycerol kinase
|
|
chr19_+_14544100
|
0.691
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr17_-_42201009
|
0.688
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chrY_+_2803409
|
0.685
|
NM_001145275
NM_003411
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr21_-_43430448
|
0.684
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr2_-_99771186
|
0.682
|
NM_025244
|
TSGA10
|
testis specific, 10
|
|
chr9_-_90589593
|
0.682
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr15_-_40401041
|
0.679
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr7_-_73184535
|
0.667
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr9_-_97356007
|
0.659
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chr14_+_24584321
|
0.648
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr2_-_160654736
|
0.646
|
NM_001198763
NM_001198764
NM_014880
|
CD302
|
CD302 molecule
|
|
chr7_+_150065330
|
0.645
|
|
REPIN1
|
replication initiator 1
|
|
chr11_-_119066583
|
0.645
|
NM_001145018
|
CCDC153
|
coiled-coil domain containing 153
|
|
chr10_+_69644708
|
0.642
|
|
SIRT1
|
sirtuin 1
|
|
chr9_+_135754284
|
0.641
|
NM_018956
|
C9orf9
|
chromosome 9 open reading frame 9
|
|
chr14_+_24583979
|
0.636
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr14_+_24584467
|
0.636
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr17_-_7137675
|
0.633
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr12_-_122107559
|
0.623
|
NM_173855
|
MORN3
|
MORN repeat containing 3
|
|
chr19_-_10764449
|
0.621
|
|
LOC147727
|
uncharacterized LOC147727
|
|
chr17_-_7137722
|
0.618
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr3_-_45883562
|
0.614
|
NM_020347
|
LZTFL1
|
leucine zipper transcription factor-like 1
|
|
chr14_+_45431333
|
0.612
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr1_+_183605207
|
0.612
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr17_-_7137580
|
0.605
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr9_-_99382073
|
0.600
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr6_+_43612766
|
0.598
|
NM_001193341
NM_152732
|
RSPH9
|
radial spoke head 9 homolog (Chlamydomonas)
|
|
chr14_+_24583905
|
0.594
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr20_+_33146500
|
0.594
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr5_+_139927182
|
0.587
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr11_-_108093328
|
0.587
|
NM_002519
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus
|
|
chr18_+_9136742
|
0.581
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr19_+_14544216
|
0.578
|
|
PKN1
|
protein kinase N1
|
|
chr10_-_123357503
|
0.571
|
NM_000141
NM_001144917
NM_001144918
NM_001144919
NM_022970
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr15_-_50978921
|
0.567
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr12_+_20963637
|
0.567
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr15_+_44084039
|
0.565
|
NM_001018108
NM_001199875
NM_001199876
|
SERF2
|
small EDRK-rich factor 2
|
|
chr5_-_126366068
|
0.563
|
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr5_-_80597362
|
0.561
|
|
LOC100131067
|
uncharacterized LOC100131067
|
|
chr22_-_41593466
|
0.560
|
|
|
|
|
chr22_+_35777051
|
0.559
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr5_-_100238870
|
0.552
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr14_+_55738102
|
0.545
|
|
FBXO34
|
F-box protein 34
|
|
chr6_+_87865261
|
0.536
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr19_-_11545900
|
0.527
|
NM_145045
|
CCDC151
|
coiled-coil domain containing 151
|
|
chr6_-_90121966
|
0.526
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr9_+_135753711
|
0.525
|
|
C9orf9
|
chromosome 9 open reading frame 9
|
|
chr16_+_4784320
|
0.524
|
|
C16orf71
|
chromosome 16 open reading frame 71
|
|
chrX_-_13835187
|
0.519
|
|
GPM6B
|
glycoprotein M6B
|
|
chr15_-_50978726
|
0.518
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr10_+_76586299
|
0.516
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chrY_+_2803111
|
0.512
|
NM_001145276
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr17_+_42977141
|
0.511
|
NM_213607
|
CCDC103
|
coiled-coil domain containing 103
|
|
chrX_+_53449804
|
0.510
|
NM_144968
NM_001031745
|
RIBC1
|
RIB43A domain with coiled-coils 1
|
|
chr11_+_101785720
|
0.510
|
NM_020802
|
KIAA1377
|
KIAA1377
|
|
chr1_+_9599546
|
0.509
|
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chrX_-_13835012
|
0.507
|
|
GPM6B
|
glycoprotein M6B
|
|
chr20_+_32077780
|
0.504
|
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr22_+_35777087
|
0.503
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr6_+_52285804
|
0.497
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr12_-_63328663
|
0.497
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr21_+_33784751
|
0.496
|
NM_058187
|
C21orf63
|
chromosome 21 open reading frame 63
|
|
chr11_+_7618416
|
0.487
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr5_+_122424840
|
0.486
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr3_-_122512526
|
0.484
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr4_-_149363498
|
0.483
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr4_+_39184023
|
0.472
|
NM_025132
|
WDR19
|
WD repeat domain 19
|
|
chr12_-_64616032
|
0.472
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr6_-_32920812
|
0.471
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr17_-_46688279
|
0.471
|
|
HOXB7
|
homeobox B7
|
|
chr11_+_60197061
|
0.462
|
NM_023945
|
MS4A5
|
membrane-spanning 4-domains, subfamily A, member 5
|
|
chr14_-_88459459
|
0.461
|
|
GALC
|
galactosylceramidase
|
|
chr16_-_88923348
|
0.458
|
NM_000512
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr1_+_85527992
|
0.457
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr16_+_4784261
|
0.454
|
NM_139170
|
C16orf71
|
chromosome 16 open reading frame 71
|
|
chr11_+_7535213
|
0.447
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr4_+_25657434
|
0.447
|
NM_001177998
NM_006424
|
SLC34A2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr3_-_36986535
|
0.446
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr16_-_46782104
|
0.446
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr6_-_84937313
|
0.439
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr2_-_60780404
|
0.438
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr19_-_36545646
|
0.437
|
NM_152658
|
THAP8
|
THAP domain containing 8
|
|
chr11_-_8739398
|
0.436
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr7_+_23719743
|
0.430
|
NM_001127364
NM_001127365
NM_199136
|
C7orf46
|
chromosome 7 open reading frame 46
|
|
chr4_+_37455551
|
0.424
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr22_+_35695872
|
0.422
|
|
TOM1
|
target of myb1 (chicken)
|
|
chrX_+_101975641
|
0.422
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr14_+_93651295
|
0.419
|
NM_015676
NM_001098621
|
C14orf109
|
chromosome 14 open reading frame 109
|
|
chr17_+_14204326
|
0.419
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr2_-_197036301
|
0.418
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr9_-_124990949
|
0.416
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr4_-_8442427
|
0.416
|
NM_001101667
NM_003501
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr15_-_40398573
|
0.410
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr9_+_6757919
|
0.406
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr5_-_121412805
|
0.405
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr7_-_36429695
|
0.405
|
NM_001100425
|
KIAA0895
|
KIAA0895
|
|
chr11_-_2016743
|
0.405
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chrX_-_34675366
|
0.403
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr2_-_178257380
|
0.403
|
|
LOC100130691
|
uncharacterized LOC100130691
|
|
chr16_+_4784446
|
0.401
|
|
C16orf71
|
chromosome 16 open reading frame 71
|
|
chrX_-_8700170
|
0.399
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chrY_+_15016775
|
0.397
|
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr16_+_699277
|
0.394
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr6_-_31745048
|
0.393
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr17_+_4402128
|
0.392
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr7_+_150068252
|
0.392
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr17_+_79609298
|
0.392
|
NM_031945
|
TSPAN10
|
tetraspanin 10
|
|
chr12_+_12764755
|
0.387
|
NM_001310
|
CREBL2
|
cAMP responsive element binding protein-like 2
|
|
chr11_-_8892909
|
0.387
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr20_-_44519629
|
0.385
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr9_-_139922595
|
0.385
|
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr2_+_120302024
|
0.385
|
NM_001029996
|
PCDP1
|
primary ciliary dyskinesia protein 1
|
|
chr12_-_65153182
|
0.384
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr1_+_154193294
|
0.384
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr15_+_44084493
|
0.383
|
|
SERF2
|
small EDRK-rich factor 2
|
|
chr17_+_73472578
|
0.383
|
|
KIAA0195
|
KIAA0195
|
|
chr11_-_6640645
|
0.381
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr1_+_151483861
|
0.381
|
NM_020770
|
CGN
|
cingulin
|
|
chr11_+_61522860
|
0.379
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr9_-_139922705
|
0.377
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr4_+_36285643
|
0.376
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr6_+_43027368
|
0.375
|
NM_138343
NM_201521
|
KLC4
|
kinesin light chain 4
|
|
chr2_-_220083699
|
0.374
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr6_+_139456240
|
0.374
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr22_-_38349533
|
0.374
|
NM_032561
NM_001207062
|
C22orf23
|
chromosome 22 open reading frame 23
|
|
chr5_-_137368682
|
0.373
|
|
FAM13B
|
family with sequence similarity 13, member B
|
|
chr6_+_32821924
|
0.373
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr4_+_175204827
|
0.372
|
NM_001040157
|
CEP44
|
centrosomal protein 44kDa
|
|
chr11_+_120894756
|
0.371
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chr17_-_57184116
|
0.369
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr4_+_106068009
|
0.369
|
|
TET2
|
tet methylcytosine dioxygenase 2
|
|
chr13_+_48877950
|
0.367
|
|
RB1
|
retinoblastoma 1
|
|
chr20_+_47538211
|
0.366
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr6_+_89791510
|
0.366
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr6_-_90121911
|
0.364
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr1_-_204329043
|
0.363
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr6_+_127587984
|
0.362
|
|
RNF146
|
ring finger protein 146
|
|
chr17_+_55333843
|
0.362
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr14_-_45431090
|
0.360
|
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr2_-_197036262
|
0.360
|
|
STK17B
|
serine/threonine kinase 17b
|
|
chr16_+_28875181
|
0.357
|
NM_001145812
NM_015503
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr12_+_38710556
|
0.356
|
NM_001013620
|
ALG10B
|
asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog B (yeast)
|
|
chr3_+_113616317
|
0.356
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr2_-_99224868
|
0.355
|
NM_001008215
|
COA5
|
cytochrome C oxidase assembly factor 5
|
|
chr2_+_46926049
|
0.355
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr11_+_67776016
|
0.354
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr14_+_74035762
|
0.354
|
NM_006821
|
ACOT2
|
acyl-CoA thioesterase 2
|
|
chr1_-_202896368
|
0.354
|
NM_021633
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr1_+_40839368
|
0.350
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr7_+_150065968
|
0.347
|
|
REPIN1
|
replication initiator 1
|
|
chr14_+_55738020
|
0.347
|
NM_017943
|
FBXO34
|
F-box protein 34
|
|
chr12_-_7245048
|
0.346
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr22_+_45809541
|
0.346
|
NM_015653
|
RIBC2
|
RIB43A domain with coiled-coils 2
|
|
chr21_-_47743753
|
0.345
|
NM_058180
|
C21orf58
|
chromosome 21 open reading frame 58
|
|
chr3_-_107941168
|
0.344
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr3_-_187463474
|
0.344
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr5_-_137368778
|
0.343
|
NM_001101800
NM_001101801
NM_016603
|
FAM13B
|
family with sequence similarity 13, member B
|