|
chr22_-_39190103
|
4.848
|
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr22_-_39190146
|
4.526
|
NM_005740
|
DNAL4
|
dynein, axonemal, light chain 4
|
|
chr1_+_85527992
|
3.779
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr10_+_63422718
|
3.559
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr9_+_140135738
|
3.193
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr12_-_113658826
|
3.006
|
NM_138451
|
IQCD
|
IQ motif containing D
|
|
chr3_+_129158949
|
2.987
|
NM_018262
NM_052985
NM_052989
NM_052990
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_129159075
|
2.946
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_129159150
|
2.945
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_129159138
|
2.908
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr3_+_129159089
|
2.899
|
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas)
|
|
chr9_+_140135698
|
2.833
|
NM_006088
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr4_-_186300151
|
2.796
|
NM_018409
|
LRP2BP
|
LRP2 binding protein
|
|
chr8_+_99076749
|
2.635
|
NM_001170806
NM_173549
|
C8orf47
|
chromosome 8 open reading frame 47
|
|
chr9_+_140135742
|
2.548
|
|
TUBB4B
|
tubulin, beta 4B class IVb
|
|
chr7_-_138363789
|
2.495
|
NM_001139456
|
SVOPL
|
SVOP-like
|
|
chr3_-_121553907
|
1.761
|
NM_001023570
NM_001023571
|
IQCB1
|
IQ motif containing B1
|
|
chr6_-_32557431
|
1.718
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr3_-_121553836
|
1.685
|
|
IQCB1
|
IQ motif containing B1
|
|
chr13_-_61989209
|
1.631
|
|
PCDH20
|
protocadherin 20
|
|
chr2_-_154335181
|
1.553
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr5_+_140018998
|
1.525
|
NM_018502
|
TMCO6
|
transmembrane and coiled-coil domains 6
|
|
chr11_+_73661363
|
1.477
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr15_+_43809805
|
1.474
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr6_+_135502477
|
1.430
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr12_+_21168629
|
1.408
|
NM_001009562
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional)
|
|
chr6_-_167040700
|
1.360
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr1_-_149858114
|
1.317
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr2_+_219135114
|
1.312
|
NM_001077399
NM_015488
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr15_-_37391893
|
1.295
|
|
MEIS2
|
Meis homeobox 2
|
|
chr4_-_69886112
|
1.292
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr17_+_58755158
|
1.272
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr13_-_61989654
|
1.267
|
NM_022843
|
PCDH20
|
protocadherin 20
|
|
chr3_-_112564789
|
1.253
|
NM_001008784
NM_001199215
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr1_+_151630710
|
1.227
|
|
SNX27
|
sorting nexin family member 27
|
|
chr18_+_44526786
|
1.221
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr2_+_12856813
|
1.197
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr5_+_43602738
|
1.184
|
NM_012343
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr13_+_97928414
|
1.184
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr16_+_30907927
|
1.176
|
NM_001142544
NM_001330
|
CTF1
|
cardiotrophin 1
|
|
chr13_+_36050885
|
1.156
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr7_+_29603388
|
1.141
|
NM_175887
|
PRR15
|
proline rich 15
|
|
chr10_+_69869249
|
1.126
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr14_+_76127550
|
1.070
|
NM_015072
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr18_-_53177999
|
1.058
|
NM_001243231
|
TCF4
|
transcription factor 4
|
|
chr14_+_24584321
|
1.031
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr13_+_43355685
|
1.030
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr2_+_183943468
|
1.005
|
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr5_+_43603216
|
1.000
|
NM_182977
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr5_+_43603312
|
0.990
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr16_+_15596122
|
0.960
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr9_-_99382073
|
0.947
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr6_+_135502521
|
0.921
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr15_-_93616308
|
0.920
|
|
RGMA
|
RGM domain family, member A
|
|
chr3_+_63638343
|
0.907
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr1_+_60280462
|
0.905
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr2_+_183943286
|
0.892
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr17_+_1619816
|
0.887
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr19_-_50528683
|
0.865
|
NM_001025778
NM_016440
|
VRK3
|
vaccinia related kinase 3
|
|
chr20_+_57022460
|
0.857
|
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr1_-_217250230
|
0.855
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr15_-_93616358
|
0.842
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr14_+_76127656
|
0.829
|
|
TTLL5
|
tubulin tyrosine ligase-like family, member 5
|
|
chr15_-_37393364
|
0.820
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr19_-_41870001
|
0.820
|
NM_030578
|
B9D2
|
B9 protein domain 2
|
|
chr19_+_49375677
|
0.819
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr9_-_37465384
|
0.812
|
NM_014872
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr4_-_84035888
|
0.807
|
|
PLAC8
|
placenta-specific 8
|
|
chr19_+_49375679
|
0.806
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr17_+_21030236
|
0.789
|
NM_015510
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B
|
|
chr5_+_43603287
|
0.787
|
|
NNT
|
nicotinamide nucleotide transhydrogenase
|
|
chr2_-_219134795
|
0.782
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr2_-_219134865
|
0.769
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr19_+_49375638
|
0.769
|
NM_014330
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr4_+_77356252
|
0.763
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr16_+_78062985
|
0.762
|
NM_001244755
|
CLEC3A
|
C-type lectin domain family 3, member A
|
|
chr1_+_118148506
|
0.761
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr18_-_59854179
|
0.751
|
|
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N
|
|
chr19_-_50528627
|
0.748
|
|
VRK3
|
vaccinia related kinase 3
|
|
chr17_+_17876126
|
0.736
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr2_-_219134846
|
0.714
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr18_-_59854288
|
0.713
|
NM_012327
NM_176787
|
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N
|
|
chr16_+_67927170
|
0.712
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr2_+_183943511
|
0.711
|
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr1_+_104159953
|
0.703
|
NM_000699
|
AMY2A
|
amylase, alpha 2A (pancreatic)
|
|
chr10_+_99498233
|
0.689
|
NM_001002261
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr6_+_27114860
|
0.682
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chrX_-_10645726
|
0.675
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr17_-_41977966
|
0.672
|
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr10_+_76586299
|
0.650
|
NM_012330
|
KAT6B
|
K(lysine) acetyltransferase 6B
|
|
chr7_-_77325539
|
0.630
|
|
|
|
|
chr14_-_93673360
|
0.629
|
NM_032490
|
C14orf142
|
chromosome 14 open reading frame 142
|
|
chr16_+_19078958
|
0.627
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr6_-_167040659
|
0.619
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr8_+_18079176
|
0.619
|
NM_001160174
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr2_-_135782246
|
0.615
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr12_+_41831484
|
0.614
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr16_-_3493489
|
0.608
|
NM_152457
|
ZNF597
|
zinc finger protein 597
|
|
chr15_-_77712436
|
0.608
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr14_+_24584009
|
0.605
|
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr11_-_85397215
|
0.604
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr6_-_31239489
|
0.603
|
|
HLA-B
HLA-C
|
major histocompatibility complex, class I, B
major histocompatibility complex, class I, C
|
|
chr6_+_16129285
|
0.596
|
NM_013262
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr14_+_24583979
|
0.592
|
NM_025230
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_+_29910308
|
0.591
|
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr12_+_55248298
|
0.587
|
NM_058173
|
MUCL1
|
mucin-like 1
|
|
chr6_+_26217145
|
0.585
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr18_-_59854215
|
0.582
|
|
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N
|
|
chr1_+_149858466
|
0.581
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr1_-_38397417
|
0.577
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr6_-_46620229
|
0.577
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chrX_+_9431314
|
0.573
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr16_+_19078916
|
0.558
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr3_+_181429711
|
0.555
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr19_+_39882018
|
0.554
|
|
MED29
|
mediator complex subunit 29
|
|
chrY_+_14813159
|
0.553
|
NM_004654
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked
|
|
chr5_+_172483285
|
0.550
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr9_-_3223007
|
0.545
|
|
|
|
|
chr6_-_26271588
|
0.545
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr19_+_9434447
|
0.543
|
NM_001202406
NM_001202407
NM_001202408
NM_001202409
NM_001202410
NM_001202411
NM_032497
|
ZNF559
|
zinc finger protein 559
|
|
chr4_-_84035908
|
0.540
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr6_+_31430955
|
0.539
|
|
HCP5
|
HLA complex P5 (non-protein coding)
|
|
chr7_+_77325729
|
0.537
|
NM_198467
|
RSBN1L
|
round spermatid basic protein 1-like
|
|
chrX_+_69674914
|
0.534
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr14_+_64971504
|
0.533
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr14_+_24583905
|
0.530
|
NM_001163484
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_167040506
|
0.530
|
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr12_-_15091441
|
0.529
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr1_+_164528586
|
0.528
|
NM_001204961
NM_001204963
NM_002585
|
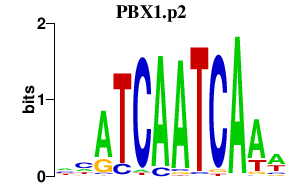
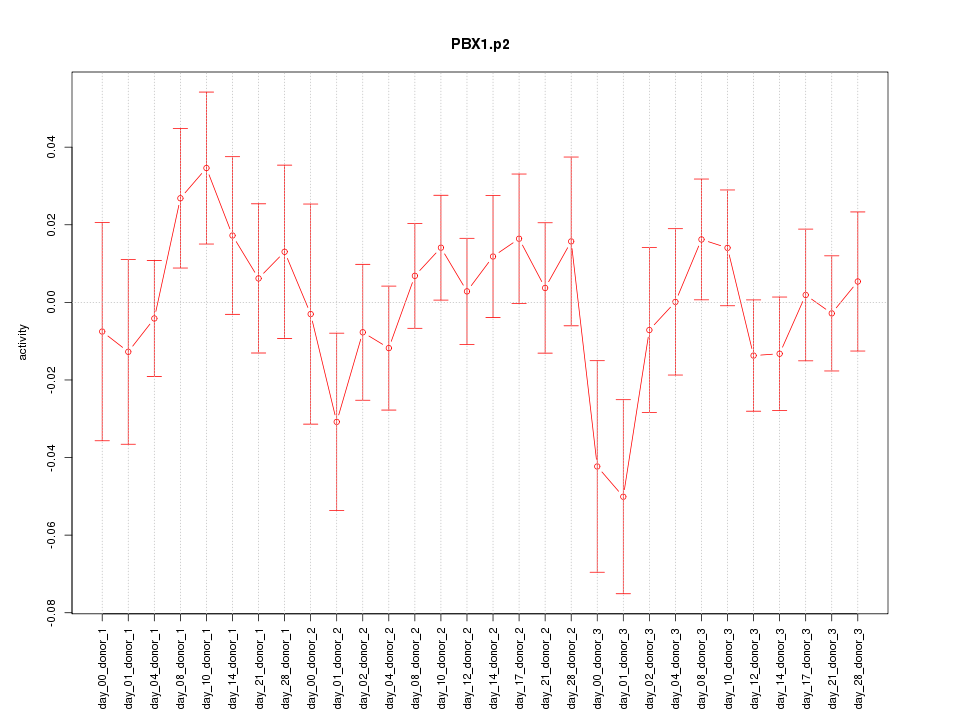
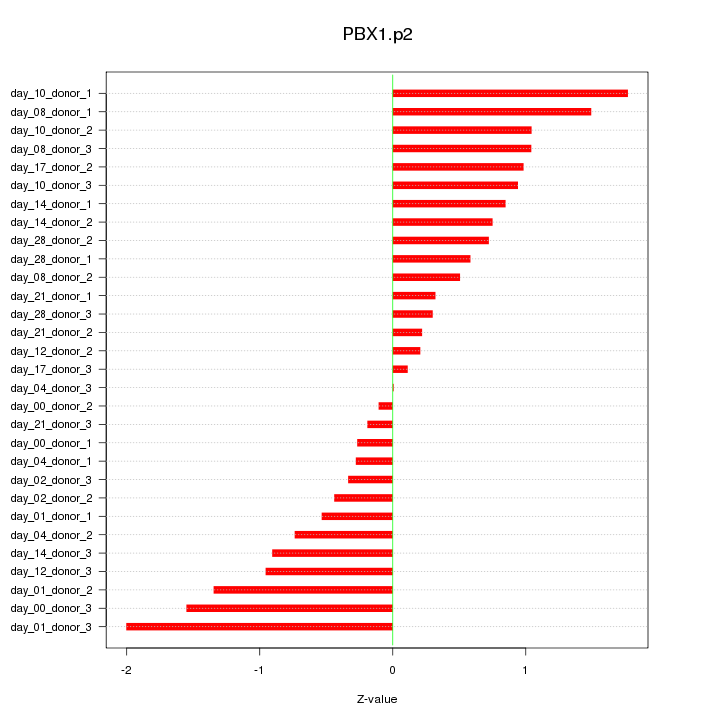
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr13_+_97928196
|
0.527
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr5_-_75013209
|
0.521
|
NM_001099271
|
POC5
|
POC5 centriolar protein homolog (Chlamydomonas)
|
|
chr5_+_140186658
|
0.520
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr14_+_23938883
|
0.513
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr3_-_148804122
|
0.507
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr16_+_57769667
|
0.504
|
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr17_+_57408907
|
0.503
|
NM_001005404
|
YPEL2
|
yippee-like 2 (Drosophila)
|
|
chr1_+_12806162
|
0.495
|
NM_152290
|
C1orf158
|
chromosome 1 open reading frame 158
|
|
chr1_+_118148668
|
0.494
|
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr1_+_6845381
|
0.490
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr8_-_79717540
|
0.485
|
NM_000880
NM_001199886
NM_001199887
NM_001199888
|
IL7
|
interleukin 7
|
|
chr3_-_148939784
|
0.480
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr16_+_82068830
|
0.477
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr18_+_32073343
|
0.476
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr19_+_39881950
|
0.472
|
NM_017592
|
MED29
|
mediator complex subunit 29
|
|
chr3_-_149510609
|
0.469
|
NM_001144960
|
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1
|
|
chr6_-_31324898
|
0.467
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr14_-_75530716
|
0.467
|
NM_001107
NM_203488
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type
|
|
chr1_+_14075917
|
0.466
|
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr6_-_42713822
|
0.459
|
|
TBCC
|
tubulin folding cofactor C
|
|
chr10_-_72142344
|
0.458
|
NM_018205
NM_018239
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr15_-_86338017
|
0.453
|
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr1_+_104292439
|
0.451
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr5_+_142149970
|
0.447
|
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr14_+_23938902
|
0.443
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr1_+_14075861
|
0.430
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr4_+_69681709
|
0.429
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr2_-_174829951
|
0.426
|
|
SP3
|
Sp3 transcription factor
|
|
chr1_+_151227209
|
0.424
|
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|
|
chr1_+_226250413
|
0.423
|
|
H3F3A
|
H3 histone, family 3A
|
|
chr14_-_58764805
|
0.423
|
|
FLJ31306
|
uncharacterized LOC379025
|
|
chr8_-_38008332
|
0.418
|
NM_000349
|
STAR
|
steroidogenic acute regulatory protein
|
|
chr7_-_95225802
|
0.411
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr6_+_135502445
|
0.409
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr8_+_107738239
|
0.408
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr5_+_67511601
|
0.406
|
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr14_-_74551073
|
0.403
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr2_+_200323088
|
0.403
|
|
|
|
|
chr7_+_21467572
|
0.401
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr6_+_31783280
|
0.401
|
NM_005345
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr14_-_91526912
|
0.399
|
NM_004755
NM_182398
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5
|
|
chr1_-_183560010
|
0.394
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr16_+_15031299
|
0.394
|
NM_006985
|
NPIP
|
nuclear pore complex interacting protein
|
|
chr16_+_639668
|
0.390
|
NM_001172664
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr1_-_28241233
|
0.389
|
NM_002946
|
RPA2
|
replication protein A2, 32kDa
|
|
chr22_+_29138018
|
0.388
|
NM_172002
|
HSCB
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli)
|
|
chr6_-_33041266
|
0.385
|
NM_033554
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr6_-_31324955
|
0.383
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr2_+_24807345
|
0.383
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr13_+_24734788
|
0.381
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr5_-_89705477
|
0.380
|
NM_004365
|
CETN3
|
centrin, EF-hand protein, 3
|
|
chr16_+_82068910
|
0.377
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr6_+_31783314
|
0.371
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr1_-_228645517
|
0.371
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr19_+_41770273
|
0.366
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr7_+_105603656
|
0.365
|
NM_152750
|
CDHR3
|
cadherin-related family member 3
|
|
chr17_-_73511626
|
0.365
|
NM_020753
|
CASKIN2
|
CASK interacting protein 2
|
|
chr9_-_124989755
|
0.364
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr12_+_121124958
|
0.362
|
|
MLEC
|
malectin
|
|
chr1_+_15990300
|
0.362
|
|
|
|
|
chr6_+_16129321
|
0.358
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr9_-_84304378
|
0.353
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr16_+_19079220
|
0.345
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr1_+_161136180
|
0.343
|
NM_000309
NM_001122764
|
PPOX
|
protoporphyrinogen oxidase
|
|
chr6_-_26235160
|
0.341
|
NM_005320
|
HIST1H1D
|
histone cluster 1, H1d
|
|
chr1_+_151227188
|
0.340
|
NM_002810
|
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4
|
|
chr19_+_41430123
|
0.338
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr9_+_1050333
|
0.338
|
NM_006557
NM_001130865
|
DMRT2
|
doublesex and mab-3 related transcription factor 2
|
|
chr12_+_58138663
|
0.337
|
NM_005981
|
TSPAN31
|
tetraspanin 31
|
|
chr3_-_52931341
|
0.337
|
|
TMEM110
|
transmembrane protein 110
|
|
chr8_+_98788014
|
0.334
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|