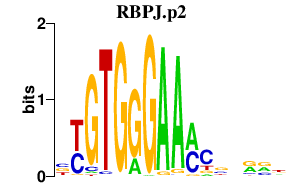
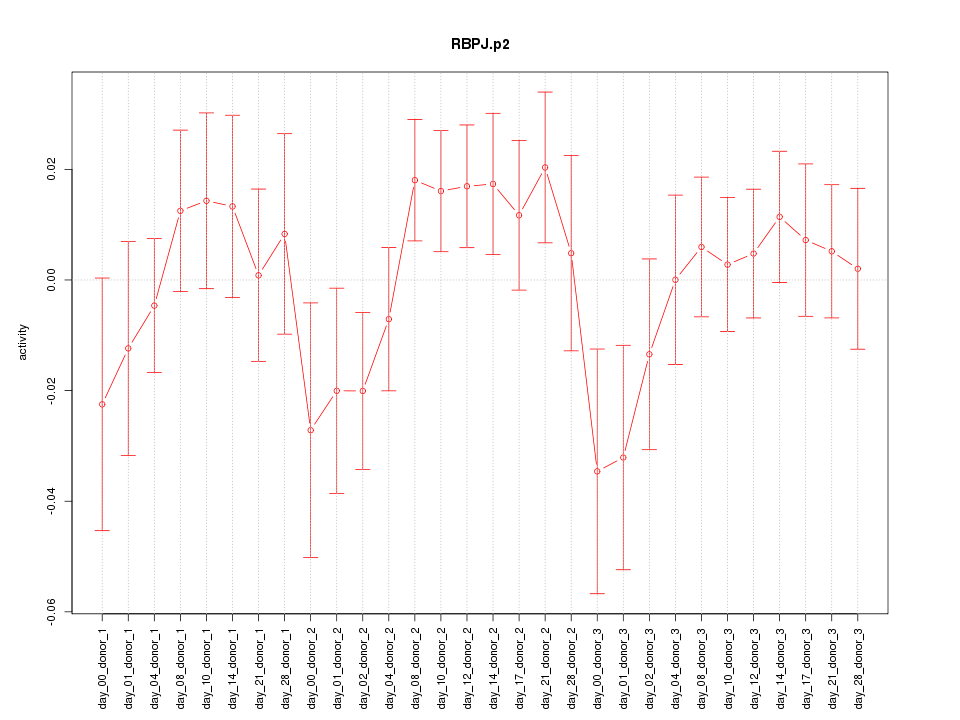
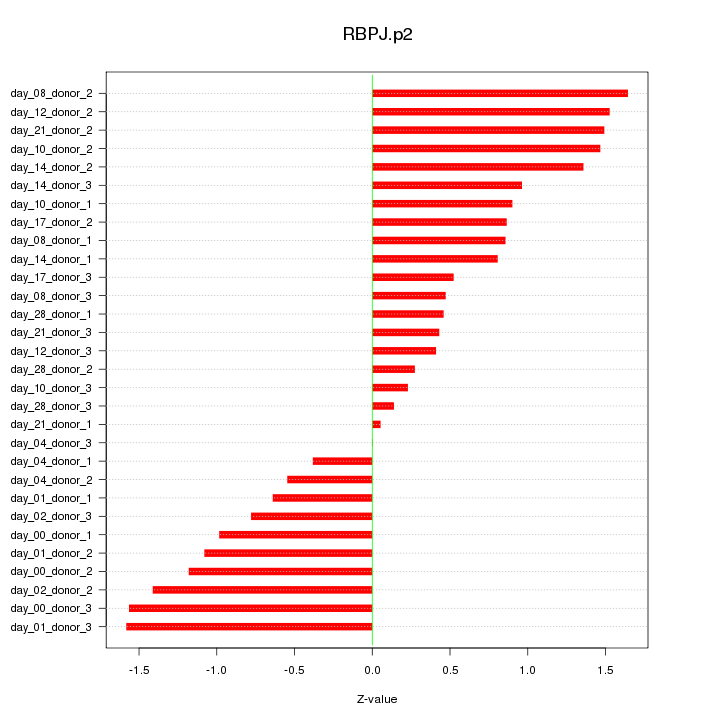
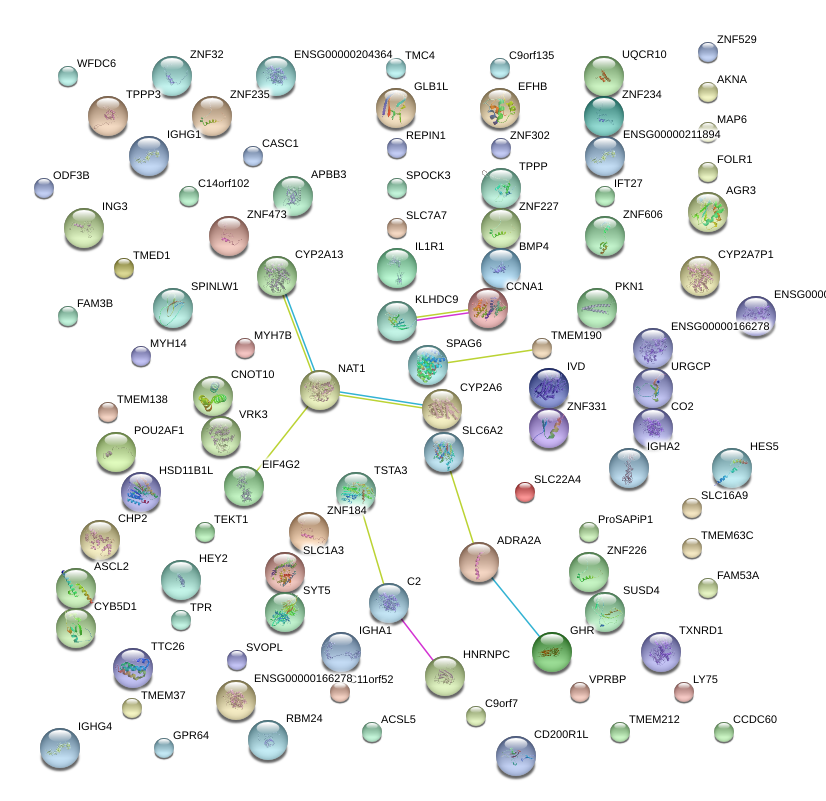
|
chr9_+_72435730
|
10.204
|
NM_001010940
|
C9orf135
|
chromosome 9 open reading frame 135
|
|
chr3_-_19975671
|
4.258
|
NM_144715
|
EFHB
|
EF-hand domain family, member B
|
|
chr16_+_23765947
|
4.062
|
NM_022097
|
CHP2
|
calcineurin B homologous protein 2
|
|
chr20_-_3149070
|
3.492
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr19_+_55888203
|
3.436
|
NM_139172
|
TMEM190
|
transmembrane protein 190
|
|
chr7_-_16921596
|
3.283
|
NM_176813
|
AGR3
|
anterior gradient 3 homolog (Xenopus laevis)
|
|
chr6_+_126070813
|
2.600
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr3_-_112564789
|
2.571
|
NM_001008784
NM_001199215
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr5_-_693413
|
2.542
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr16_-_67427335
|
2.424
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr19_-_50528683
|
2.351
|
NM_001025778
NM_016440
|
VRK3
|
vaccinia related kinase 3
|
|
chr6_+_126070724
|
2.306
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr1_-_223537400
|
2.293
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr8_-_144699716
|
2.292
|
NM_003313
|
TSTA3
|
tissue specific transplantation antigen P35B
|
|
chr14_-_54421269
|
2.217
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr8_-_144699695
|
2.194
|
|
TSTA3
|
tissue specific transplantation antigen P35B
|
|
chr2_+_120189342
|
2.175
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr11_-_2292181
|
2.061
|
NM_005170
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr10_-_44144151
|
2.026
|
NM_001005368
|
ZNF32
|
zinc finger protein 32
|
|
chr5_+_42423811
|
2.013
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr8_-_144699621
|
1.967
|
|
TSTA3
|
tissue specific transplantation antigen P35B
|
|
chr19_-_50528627
|
1.940
|
|
VRK3
|
vaccinia related kinase 3
|
|
chr2_-_160761266
|
1.821
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr19_-_37064102
|
1.756
|
|
ZNF529
|
zinc finger protein 529
|
|
chr2_-_160761192
|
1.749
|
|
LY75-CD302
LY75
|
LY75-CD302 readthrough
lymphocyte antigen 75
|
|
chr11_+_111789600
|
1.740
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr19_+_54024176
|
1.684
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr5_+_131630065
|
1.670
|
NM_003059
|
SLC22A4
|
solute carrier family 22 (organic cation/ergothioneine transporter), member 4
|
|
chr11_-_75379651
|
1.656
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr6_+_126070738
|
1.598
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr14_-_23285011
|
1.583
|
NM_001126105
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr13_+_37006628
|
1.570
|
|
CCNA1
|
cyclin A1
|
|
chr7_+_138818489
|
1.566
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr10_+_22634373
|
1.561
|
NM_001253854
NM_001253855
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr3_-_51533906
|
1.552
|
|
VPRBP
|
Vpr (HIV-1) binding protein
|
|
chr19_-_50528576
|
1.544
|
|
VRK3
|
vaccinia related kinase 3
|
|
chr3_+_171561138
|
1.466
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr11_-_2291809
|
1.449
|
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr19_+_5681016
|
1.444
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr13_+_37006408
|
1.428
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr19_+_58816714
|
1.394
|
|
ERVK3-1
|
endogenous retrovirus group K3, member 1
|
|
chr1_+_161068150
|
1.391
|
NM_001007255
NM_152366
|
KLHDC9
|
kelch domain containing 9
|
|
chr19_-_55690717
|
1.379
|
|
SYT5
|
synaptotagmin V
|
|
chr2_+_102770401
|
1.361
|
NM_000877
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr19_+_58816757
|
1.355
|
|
ERVK3-1
|
endogenous retrovirus group K3, member 1
|
|
chr17_-_6735007
|
1.350
|
NM_053285
|
TEKT1
|
tektin 1
|
|
chr7_+_120590816
|
1.349
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr19_+_14544100
|
1.332
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr11_+_71903172
|
1.329
|
NM_016729
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr19_+_44645728
|
1.325
|
|
ZNF234
|
zinc finger protein 234
|
|
chr15_+_40697977
|
1.323
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr19_+_35168558
|
1.308
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr8_+_18067614
|
1.302
|
NM_000662
NM_001160170
NM_001160171
NM_001160172
NM_001160173
NM_001160175
NM_001160176
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr15_+_40698000
|
1.293
|
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr22_-_50970542
|
1.253
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr2_-_220110116
|
1.246
|
NM_024506
|
GLB1L
|
galactosidase, beta 1-like
|
|
chr16_+_84178864
|
1.237
|
NM_178452
|
DNAAF1
|
dynein, axonemal, assembly factor 1
|
|
chr14_+_77648127
|
1.228
|
|
TMEM63C
|
transmembrane protein 63C
|
|
chr10_+_114154522
|
1.226
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr3_+_32726659
|
1.221
|
NM_015442
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10
|
|
chr10_-_61469322
|
1.215
|
NM_194298
|
LOC100129721
SLC16A9
|
uncharacterized LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr6_+_17281773
|
1.172
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chrX_-_19140581
|
1.171
|
NM_001079858
NM_001079859
NM_001079860
NM_001184833
NM_001184834
NM_001184835
NM_001184836
NM_001184837
NM_005756
|
GPR64
|
G protein-coupled receptor 64
|
|
chr5_-_139943829
|
1.156
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr11_+_94245694
|
1.155
|
NM_001190462
|
LOC643037
|
uncharacterized LOC643037
|
|
chr6_+_32811895
|
1.150
|
|
|
|
|
chr19_+_50529211
|
1.144
|
NM_001006656
NM_015428
|
ZNF473
|
zinc finger protein 473
|
|
chr19_-_54676813
|
1.140
|
NM_001145303
NM_144686
|
TMC4
|
transmembrane channel-like 4
|
|
chr14_-_106926393
|
1.134
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_-_44144266
|
1.104
|
NM_006973
|
ZNF32
|
zinc finger protein 32
|
|
chr19_+_50706868
|
1.096
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr17_+_7761310
|
1.057
|
|
CYB5D1
|
cytochrome b5 domain containing 1
|
|
chr21_+_42676176
|
1.052
|
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr14_-_90798253
|
1.051
|
NM_017970
|
C14orf102
|
chromosome 14 open reading frame 102
|
|
chr1_-_2461683
|
1.051
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr11_-_111250156
|
1.042
|
NM_006235
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr5_+_36608430
|
1.032
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr14_+_77648082
|
1.030
|
NM_020431
|
TMEM63C
|
transmembrane protein 63C
|
|
chr19_+_44716683
|
1.015
|
NM_182490
|
ZNF227
|
zinc finger protein 227
|
|
chr19_+_41594367
|
1.012
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr7_-_138363789
|
1.006
|
NM_001139456
|
SVOPL
|
SVOP-like
|
|
chr19_+_5681139
|
0.994
|
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr11_+_61129407
|
0.977
|
NM_016464
|
TMEM138
|
transmembrane protein 138
|
|
chr7_+_150065968
|
0.973
|
|
REPIN1
|
replication initiator 1
|
|
chr22_-_37164073
|
0.970
|
NM_001177702
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr4_-_1685717
|
0.965
|
NM_001013622
NM_001174070
|
FAM53A
|
family with sequence similarity 53, member A
|
|
chr19_-_44809126
|
0.964
|
NM_004234
|
ZNF235
|
zinc finger protein 235
|
|
chr6_-_27440459
|
0.951
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr19_+_14544216
|
0.945
|
|
PKN1
|
protein kinase N1
|
|
chr20_-_44175995
|
0.945
|
NM_001198986
NM_020398
|
SPINLW1-WFDC6
SPINLW1
|
SPINLW1-WFDC6 readthrough
serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin)
|
|
chr6_+_31895201
|
0.944
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr22_+_30163351
|
0.944
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr1_-_186344383
|
0.914
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr9_-_117156684
|
0.913
|
NM_030767
|
AKNA
|
AT-hook transcription factor
|
|
chr9_+_136325086
|
0.910
|
NM_001135775
NM_001242369
NM_001242370
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr12_-_25348031
|
0.907
|
NM_001082972
NM_001082973
NM_001204101
NM_001204102
NM_018272
|
CASC1
|
cancer susceptibility candidate 1
|
|
chr19_-_10946866
|
0.898
|
NM_006858
|
TMED1
|
transmembrane emp24 protein transport domain containing 1
|
|
chr12_+_104609501
|
0.895
|
NM_001093771
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr11_-_111249902
|
0.886
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr4_-_168155732
|
0.886
|
NM_001040159
NM_001204352
NM_001204354
NM_001204355
NM_001204356
NM_001204358
NM_001251967
NM_016950
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr12_+_119772516
|
0.873
|
NM_178499
|
CCDC60
|
coiled-coil domain containing 60
|
|
chr19_-_58514713
|
0.868
|
NM_025027
|
ZNF606
|
zinc finger protein 606
|
|
chr3_+_49027281
|
0.867
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr5_-_179498598
|
0.865
|
|
RNF130
|
ring finger protein 130
|
|
chr1_-_186344389
|
0.859
|
NM_003292
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr18_-_47814024
|
0.856
|
|
CXXC1
|
CXXC finger protein 1
|
|
chr8_+_96145968
|
0.856
|
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2
|
|
chr5_-_149792276
|
0.855
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr9_-_139923214
|
0.849
|
NM_212533
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr4_-_168155576
|
0.837
|
NM_001204353
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr16_+_23313590
|
0.836
|
NM_000336
|
SCNN1B
|
sodium channel, nonvoltage-gated 1, beta
|
|
chr7_+_150065859
|
0.836
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr11_+_61129831
|
0.831
|
|
TMEM138
|
transmembrane protein 138
|
|
chr18_-_47814005
|
0.831
|
|
CXXC1
|
CXXC finger protein 1
|
|
chr2_+_233562031
|
0.831
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr18_-_47814187
|
0.830
|
|
CXXC1
|
CXXC finger protein 1
|
|
chrX_-_15620191
|
0.824
|
NM_021804
|
ACE2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr11_+_827584
|
0.818
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr12_+_6561176
|
0.813
|
NM_018009
|
TAPBPL
|
TAP binding protein-like
|
|
chr14_-_92302612
|
0.811
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr5_+_140213968
|
0.805
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chrX_-_112084006
|
0.804
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr10_+_114133775
|
0.802
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr4_+_130014828
|
0.799
|
NM_173487
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr11_-_75379414
|
0.797
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chr1_-_186344330
|
0.786
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr9_-_85678042
|
0.777
|
NM_152573
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr13_+_24144402
|
0.772
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr2_+_233561997
|
0.771
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr19_+_9434447
|
0.770
|
NM_001202406
NM_001202407
NM_001202408
NM_001202409
NM_001202410
NM_001202411
NM_032497
|
ZNF559
|
zinc finger protein 559
|
|
chr15_+_40697685
|
0.770
|
NM_001159508
NM_002225
|
IVD
|
isovaleryl-CoA dehydrogenase
|
|
chr6_-_32805819
|
0.761
|
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr4_-_7044664
|
0.761
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chr1_-_186344241
|
0.761
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr12_+_12224401
|
0.754
|
NM_030766
NM_138722
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator)
|
|
chr7_-_16844606
|
0.750
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr15_-_49255640
|
0.749
|
NM_203349
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4
|
|
chr12_-_59314069
|
0.747
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr17_+_7608413
|
0.745
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr1_-_23810627
|
0.740
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr12_-_59314297
|
0.736
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr19_-_44143943
|
0.734
|
NM_145296
|
CADM4
|
cell adhesion molecule 4
|
|
chr8_+_96146097
|
0.732
|
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2
|
|
chr14_-_61115906
|
0.721
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr15_+_59730272
|
0.715
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr19_+_47778099
|
0.712
|
NM_178511
|
PRR24
|
proline rich 24
|
|
chr19_+_37569307
|
0.705
|
NM_144689
|
ZNF420
|
zinc finger protein 420
|
|
chr15_-_43622687
|
0.694
|
NM_014793
|
LCMT2
|
leucine carboxyl methyltransferase 2
|
|
chr19_+_9434854
|
0.691
|
NM_001172650
NM_001202412
|
ZNF559-ZNF177
ZNF559
|
ZNF559-ZNF177 readthrough
zinc finger protein 559
|
|
chr19_+_50529276
|
0.688
|
|
ZNF473
|
zinc finger protein 473
|
|
chr19_-_40596781
|
0.687
|
NM_001010880
NM_001142577
NM_001142578
NM_001142579
|
ZNF780A
|
zinc finger protein 780A
|
|
chr4_-_84256012
|
0.687
|
NM_001098540
|
HPSE
|
heparanase
|
|
chr11_-_61129273
|
0.685
|
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr16_+_640172
|
0.685
|
NM_021168
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr22_+_31003069
|
0.683
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chrX_-_151999259
|
0.683
|
NM_004344
|
CETN2
|
centrin, EF-hand protein, 2
|
|
chr3_+_99357439
|
0.682
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr14_+_57735605
|
0.682
|
NM_018229
|
MUDENG
|
MU-2/AP1M2 domain containing, death-inducing
|
|
chr15_-_65503786
|
0.680
|
NM_003613
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase
|
|
chr7_+_102987970
|
0.679
|
NM_001204453
NM_002803
|
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2
|
|
chr17_-_80231240
|
0.673
|
|
CSNK1D
|
casein kinase 1, delta
|
|
chr20_-_1165116
|
0.670
|
NM_018354
|
TMEM74B
|
transmembrane protein 74B
|
|
chr11_+_61248584
|
0.669
|
NM_001170753
NM_145017
|
PPP1R32
|
protein phosphatase 1, regulatory subunit 32
|
|
chr1_-_60392324
|
0.667
|
NM_000775
|
CYP2J2
|
cytochrome P450, family 2, subfamily J, polypeptide 2
|
|
chr22_-_39268169
|
0.664
|
|
CBX6
|
chromobox homolog 6
|
|
chr9_-_130712866
|
0.664
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr1_-_935352
|
0.663
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr6_-_32812189
|
0.658
|
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7)
|
|
chr14_-_105261850
|
0.652
|
|
AKT1
|
v-akt murine thymoma viral oncogene homolog 1
|
|
chr9_+_100174369
|
0.648
|
|
TDRD7
|
tudor domain containing 7
|
|
chrX_+_19362010
|
0.642
|
NM_000284
NM_001173454
NM_001173455
NM_001173456
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1
|
|
chr10_+_38383218
|
0.638
|
NM_001007094
NM_001178101
NM_003421
|
ZNF37A
|
zinc finger protein 37A
|
|
chr1_-_91487543
|
0.632
|
|
ZNF644
|
zinc finger protein 644
|
|
chr16_-_68482373
|
0.628
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr4_-_168155691
|
0.627
|
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr4_-_84256304
|
0.623
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr2_+_32853086
|
0.621
|
NM_001193509
NM_017735
|
TTC27
|
tetratricopeptide repeat domain 27
|
|
chr10_-_43048029
|
0.607
|
|
ZNF37BP
|
zinc finger protein 37B, pseudogene
|
|
chr17_-_62502236
|
0.603
|
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr1_-_186344801
|
0.597
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr11_+_8102691
|
0.596
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr1_+_36690038
|
0.594
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr19_+_50180408
|
0.590
|
NM_001207042
NM_001536
NM_198318
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr5_+_178450775
|
0.590
|
NM_001136116
|
ZNF879
|
zinc finger protein 879
|
|
chr3_+_49027770
|
0.575
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr8_+_123794043
|
0.571
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr19_+_35225095
|
0.571
|
|
ZNF181
|
zinc finger protein 181
|
|
chr1_+_155100304
|
0.569
|
NM_004428
NM_182685
|
EFNA1
|
ephrin-A1
|
|
chr5_-_76382905
|
0.568
|
NM_032367
|
ZBED3
|
zinc finger, BED-type containing 3
|
|
chr17_+_26698986
|
0.568
|
NM_015077
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr11_+_537521
|
0.567
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr15_-_55800431
|
0.565
|
NM_001033559
NM_001033560
NM_130810
|
DYX1C1
|
dyslexia susceptibility 1 candidate 1
|
|
chr12_+_6898705
|
0.564
|
|
CD4
|
CD4 molecule
|
|
chr12_-_7245048
|
0.563
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr19_+_50529294
|
0.562
|
|
ZNF473
|
zinc finger protein 473
|
|
chr3_+_58223228
|
0.560
|
NM_020676
|
ABHD6
|
abhydrolase domain containing 6
|
|
chr16_+_639356
|
0.558
|
NM_001172663
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr2_-_154335181
|
0.555
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr12_-_7245006
|
0.554
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr2_+_230787195
|
0.553
|
NM_174899
|
FBXO36
|
F-box protein 36
|