|
chr17_-_8113862
|
6.970
|
NM_004217
|
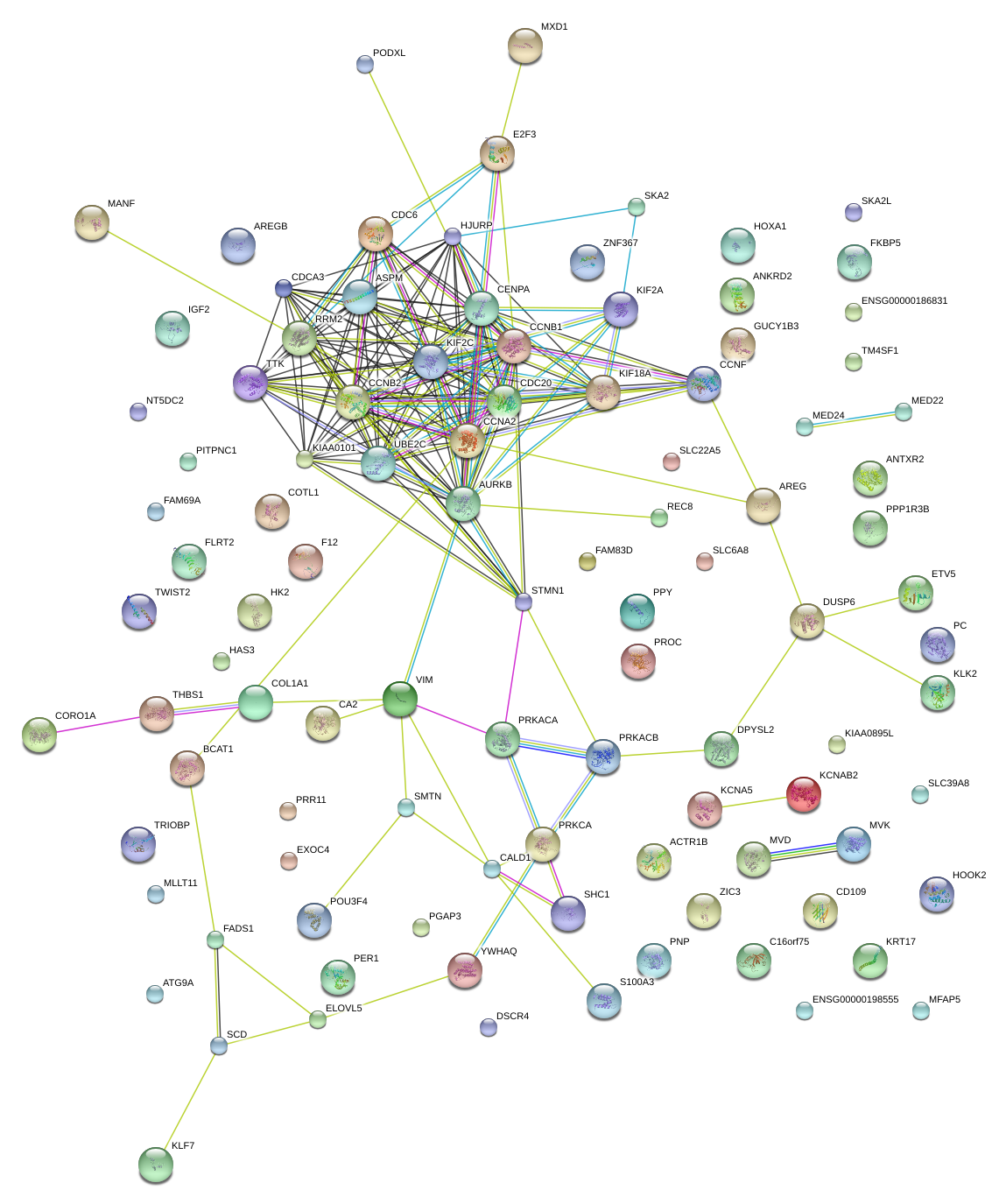
AURKB
|
aurora kinase B
|
|
chr1_+_151032866
|
6.699
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr5_+_68462948
|
6.514
|
|
CCNB1
|
cyclin B1
|
|
chr1_+_151032880
|
6.261
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr20_+_37554954
|
5.762
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr10_+_102106958
|
5.701
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr2_+_27008881
|
5.270
|
NM_001809
NM_001042426
|
CENPA
|
centromere protein A
|
|
chr3_-_52567708
|
5.248
|
NM_001134231
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr20_+_37555055
|
5.198
|
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr10_+_102106771
|
5.137
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr3_-_52567809
|
5.136
|
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr11_-_66725790
|
5.080
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr1_+_151032683
|
5.046
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_-_26232890
|
4.950
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr10_+_102107074
|
4.902
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr1_-_26232643
|
4.897
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr5_+_68462985
|
4.870
|
|
CCNB1
|
cyclin B1
|
|
chr7_-_27135524
|
4.755
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr7_+_27135672
|
4.730
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr2_+_27008912
|
4.691
|
|
CENPA
|
centromere protein A
|
|
chr14_+_24641204
|
4.534
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr1_-_26233367
|
4.518
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr1_+_43824598
|
4.383
|
NM_001255
|
CDC20
|
cell division cycle 20 homolog (S. cerevisiae)
|
|
chr17_+_57232859
|
4.373
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr7_+_134464417
|
4.208
|
|
CALD1
|
caldesmon 1
|
|
chr8_+_86376130
|
4.044
|
NM_000067
|
CA2
|
carbonic anhydrase II
|
|
chr2_+_70142303
|
3.963
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr14_+_24641084
|
3.943
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr6_+_80714389
|
3.929
|
|
TTK
|
TTK protein kinase
|
|
chr10_+_102107001
|
3.822
|
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr4_-_122744782
|
3.760
|
|
CCNA2
|
cyclin A2
|
|
chr12_-_6960406
|
3.699
|
NM_031299
|
CDCA3
|
cell division cycle associated 3
|
|
chr2_+_75061234
|
3.614
|
|
HK2
|
hexokinase 2
|
|
chr2_+_70142236
|
3.605
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr5_+_68462836
|
3.593
|
NM_031966
|
CCNB1
|
cyclin B1
|
|
chr3_-_185826748
|
3.581
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr12_-_25055228
|
3.518
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr4_-_122744978
|
3.480
|
NM_001237
|
CCNA2
|
cyclin A2
|
|
chr6_-_30655078
|
3.322
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr6_-_30654978
|
3.273
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr7_+_134464161
|
3.261
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr22_+_31477231
|
3.223
|
NM_006932
NM_134269
NM_134270
|
SMTN
|
smoothelin
|
|
chr10_+_102107256
|
3.125
|
|
|
|
|
chr1_-_154946830
|
3.098
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr4_-_80993713
|
3.087
|
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr4_+_156680125
|
3.075
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr12_-_6961044
|
3.010
|
|
CDCA3
|
cell division cycle associated 3
|
|
chr11_-_61584228
|
3.000
|
|
FADS1
|
fatty acid desaturase 1
|
|
chr6_+_74405764
|
2.944
|
|
CD109
|
CD109 molecule
|
|
chr7_+_134464372
|
2.917
|
|
CALD1
|
caldesmon 1
|
|
chr17_-_48262900
|
2.906
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr6_+_74405505
|
2.848
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr22_+_38142245
|
2.811
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr17_-_8055684
|
2.797
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr9_-_99180661
|
2.764
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr12_-_89746238
|
2.744
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr2_+_10262694
|
2.708
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr6_+_74405933
|
2.669
|
|
CD109
|
CD109 molecule
|
|
chr4_+_75310852
|
2.640
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr14_+_85996454
|
2.602
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr1_+_6052758
|
2.525
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr1_-_154946859
|
2.485
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr1_-_153521717
|
2.475
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr16_+_11439279
|
2.469
|
NM_152308
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr1_-_154946801
|
2.447
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr2_+_10262853
|
2.442
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr11_-_2162340
|
2.438
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_-_67217403
|
2.408
|
|
KIAA0895L
|
KIAA0895-like
|
|
chr20_+_44441334
|
2.406
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr20_+_44441247
|
2.406
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr16_+_2479387
|
2.404
|
NM_001761
|
CCNF
|
cyclin F
|
|
chr17_+_38444130
|
2.403
|
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr6_+_80714321
|
2.395
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr16_-_84651639
|
2.374
|
NM_021149
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr15_+_59397276
|
2.369
|
NM_004701
|
CCNB2
|
cyclin B2
|
|
chr16_+_11439311
|
2.367
|
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr15_+_39873276
|
2.362
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr6_-_53213699
|
2.335
|
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr16_+_2479417
|
2.321
|
|
CCNF
|
cyclin F
|
|
chr1_+_151032150
|
2.293
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr3_-_149086884
|
2.290
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr6_+_74405909
|
2.288
|
|
CD109
|
CD109 molecule
|
|
chr2_+_70142152
|
2.269
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr1_-_93342706
|
2.256
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr16_-_75529278
|
2.246
|
|
|
|
|
chr17_-_38210611
|
2.239
|
|
MED24
|
mediator complex subunit 24
|
|
chr6_+_20402040
|
2.155
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr2_-_208031570
|
2.143
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr1_-_197115566
|
2.122
|
NM_001206846
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chrX_+_136648313
|
2.119
|
NM_003413
|
ZIC3
|
Zic family member 3
|
|
chr17_+_65373362
|
2.111
|
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr2_-_234763145
|
2.111
|
|
HJURP
|
Holliday junction recognition protein
|
|
chr2_-_234763182
|
2.101
|
NM_018410
|
HJURP
|
Holliday junction recognition protein
|
|
chr12_-_89746244
|
2.074
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr2_-_220094354
|
2.037
|
NM_001077198
NM_024085
|
ATG9A
|
ATG9 autophagy related 9 homolog A (S. cerevisiae)
|
|
chr3_+_51422691
|
2.034
|
NM_006010
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr22_+_38142230
|
2.032
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr8_-_9009050
|
2.030
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr11_-_28129646
|
2.027
|
|
KIF18A
|
kinesin family member 18A
|
|
chr1_+_45205466
|
2.023
|
NM_006845
|
KIF2C
|
kinesin family member 2C
|
|
chr2_-_9771101
|
2.015
|
NM_006826
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr3_+_51422716
|
2.008
|
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr17_+_38444134
|
2.000
|
NM_001254
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr16_-_88729487
|
1.987
|
NM_002461
|
MVD
|
mevalonate (diphospho) decarboxylase
|
|
chr2_-_9770999
|
1.977
|
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr14_+_20937537
|
1.958
|
NM_000270
|
PNP
|
purine nucleoside phosphorylase
|
|
chr12_-_8815479
|
1.949
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr15_-_64673569
|
1.928
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr16_+_69139466
|
1.926
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr17_+_38444151
|
1.916
|
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr11_-_28129701
|
1.914
|
NM_031217
|
KIF18A
|
kinesin family member 18A
|
|
chr1_+_116654375
|
1.910
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr6_-_35696359
|
1.908
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr11_-_2152960
|
1.907
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr16_+_30194925
|
1.885
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr2_+_239756672
|
1.882
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr8_+_26435339
|
1.875
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr10_+_17271276
|
1.868
|
|
VIM
|
vimentin
|
|
chrX_+_152954965
|
1.867
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr16_-_84651519
|
1.865
|
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr11_-_28129687
|
1.863
|
|
KIF18A
|
kinesin family member 18A
|
|
chrX_+_82763268
|
1.849
|
NM_000307
|
POU3F4
|
POU class 3 homeobox 4
|
|
chr19_-_14228560
|
1.812
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr4_-_80994309
|
1.809
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr17_-_57232524
|
1.806
|
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr4_-_80994183
|
1.795
|
|
|
|
|
chr10_+_17271277
|
1.790
|
|
VIM
|
vimentin
|
|
chr12_+_110011499
|
1.789
|
NM_000431
NM_001114185
|
MVK
|
mevalonate kinase
|
|
chr10_+_17271302
|
1.788
|
|
VIM
|
vimentin
|
|
chr20_+_44441671
|
1.779
|
NM_181801
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr8_+_26435920
|
1.774
|
NM_001244604
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr12_-_89745997
|
1.769
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr5_-_176836535
|
1.767
|
NM_000505
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr4_-_103266577
|
1.764
|
NM_001135146
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr19_+_1450208
|
1.732
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr1_+_209602164
|
1.713
|
NM_001104548
|
MIR205HG
|
MIR205 host gene (non-protein coding)
|
|
chr15_+_90118740
|
1.712
|
NM_152259
|
C15orf42
|
chromosome 15 open reading frame 42
|
|
chr18_+_21718930
|
1.712
|
NM_012189
NM_138643
NM_153768
NM_153769
NM_153770
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr2_+_102314162
|
1.698
|
NM_001242560
NM_004834
NM_145686
NM_001242559
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr9_+_137218324
|
1.692
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr7_-_50861114
|
1.676
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr17_-_57232799
|
1.674
|
NM_001100595
NM_182620
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr8_+_126010748
|
1.672
|
|
SQLE
|
squalene epoxidase
|
|
chr7_-_50860665
|
1.663
|
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr1_-_221915458
|
1.663
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr16_+_838634
|
1.660
|
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr1_+_45205533
|
1.658
|
|
KIF2C
|
kinesin family member 2C
|
|
chr20_-_56285576
|
1.646
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr17_+_16945766
|
1.643
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr1_-_221915393
|
1.642
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr6_+_74405856
|
1.635
|
|
CD109
|
CD109 molecule
|
|
chr1_-_243418675
|
1.628
|
NM_001042404
NM_001042405
NM_014812
|
CEP170
|
centrosomal protein 170kDa
|
|
chr12_-_8815266
|
1.626
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr11_-_61584451
|
1.622
|
NM_013402
|
FADS1
|
fatty acid desaturase 1
|
|
chr8_+_126010719
|
1.606
|
NM_003129
|
SQLE
|
squalene epoxidase
|
|
chr16_+_30194730
|
1.600
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr1_-_221915426
|
1.597
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr11_+_13690246
|
1.591
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr7_+_100797685
|
1.587
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr6_-_27114577
|
1.581
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr16_+_838610
|
1.581
|
NM_022092
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr11_+_62186506
|
1.556
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr7_-_83824216
|
1.552
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chrX_-_15872905
|
1.540
|
|
|
|
|
chr21_-_16437125
|
1.536
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chrX_-_15872921
|
1.535
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr6_+_114178516
|
1.528
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr2_-_26205437
|
1.526
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr10_+_17271956
|
1.517
|
|
VIM
|
vimentin
|
|
chr17_-_38574042
|
1.509
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr3_+_189349215
|
1.493
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr6_+_24775640
|
1.490
|
NM_001251991
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr4_-_74864296
|
1.489
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr6_-_53213747
|
1.481
|
NM_001242828
NM_001242830
NM_001242831
NM_021814
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr15_-_34610928
|
1.478
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr4_+_166248816
|
1.475
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr5_+_54398473
|
1.475
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr7_-_41742666
|
1.463
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr20_-_52790511
|
1.460
|
NM_000782
NM_001128915
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1
|
|
chr13_+_52586516
|
1.452
|
NM_001004127
|
ALG11
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
|
|
chr1_-_6545521
|
1.435
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr7_+_77167351
|
1.432
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr10_+_17271859
|
1.430
|
|
VIM
|
vimentin
|
|
chr16_+_838645
|
1.430
|
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr3_-_139258531
|
1.429
|
NM_001130992
NM_001130993
NM_002899
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr14_-_94423766
|
1.425
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr16_-_67217825
|
1.417
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr2_-_61765345
|
1.407
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chrX_+_151999565
|
1.407
|
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chr1_+_45205558
|
1.406
|
|
KIF2C
|
kinesin family member 2C
|
|
chr9_+_80911990
|
1.405
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr6_+_160182988
|
1.381
|
NM_005891
|
ACAT2
|
acetyl-CoA acetyltransferase 2
|
|
chr3_-_87040246
|
1.381
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr15_+_41625152
|
1.366
|
|
NUSAP1
|
nucleolar and spindle associated protein 1
|
|
chr12_+_110011616
|
1.360
|
|
MVK
|
mevalonate kinase
|
|
chr1_-_236030170
|
1.358
|
|
|
|
|
chr19_+_17420352
|
1.354
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr1_+_109419601
|
1.347
|
NM_013296
|
GPSM2
|
G-protein signaling modulator 2
|
|
chr20_+_3776905
|
1.334
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr17_+_33914320
|
1.325
|
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|